
Sclerophthora macrospora virus B
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified ssRNA viruses; unclassified ssRNA positive-strand viruses
Average proteome isoelectric point is 7.67
Get precalculated fractions of proteins
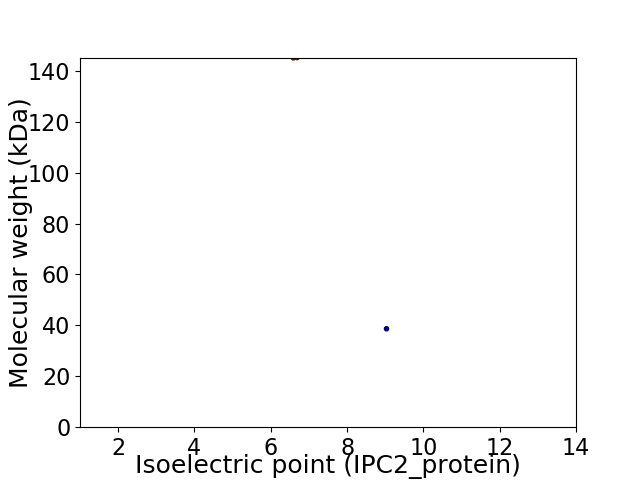
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9WTA1|Q9WTA1_9VIRU Polyprotein OS=Sclerophthora macrospora virus B OX=75914 PE=4 SV=1
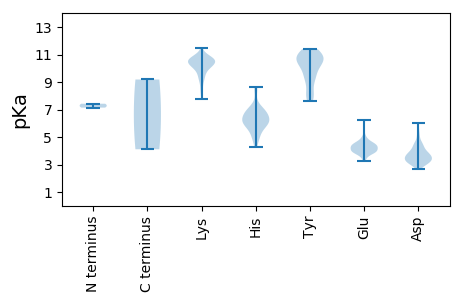
MM1 pKa = 7.42EE2 pKa = 6.21PKK4 pKa = 9.27TQVNYY9 pKa = 11.13AEE11 pKa = 4.53VVGGRR16 pKa = 11.84YY17 pKa = 8.67NRR19 pKa = 11.84PAEE22 pKa = 3.99VRR24 pKa = 11.84AMRR27 pKa = 11.84FLASNLYY34 pKa = 9.88GAGIVFDD41 pKa = 3.96ATEE44 pKa = 4.43GGAKK48 pKa = 10.09CFTSPPPHH56 pKa = 6.66PASLRR61 pKa = 11.84FCSVRR66 pKa = 11.84GMPALAVCQTQGWWASFVLKK86 pKa = 10.51SLPFPARR93 pKa = 11.84WLIFQFLNYY102 pKa = 8.76CWEE105 pKa = 4.0WVEE108 pKa = 3.96LQDD111 pKa = 4.46RR112 pKa = 11.84SAVFTATTWTLLPMAKK128 pKa = 9.43RR129 pKa = 11.84FMGPGLEE136 pKa = 4.09YY137 pKa = 10.61PEE139 pKa = 5.73IGEE142 pKa = 4.29DD143 pKa = 2.92WFDD146 pKa = 5.55RR147 pKa = 11.84IIDD150 pKa = 3.74SPRR153 pKa = 11.84YY154 pKa = 9.28RR155 pKa = 11.84GWEE158 pKa = 3.81VVPQVSYY165 pKa = 10.9LYY167 pKa = 10.68QNVGNCCLSYY177 pKa = 10.42SAHH180 pKa = 5.82EE181 pKa = 4.6KK182 pKa = 9.81IVAFVTEE189 pKa = 4.95LYY191 pKa = 10.75CRR193 pKa = 11.84FSTYY197 pKa = 10.78GIYY200 pKa = 10.67LSARR204 pKa = 11.84TLRR207 pKa = 11.84QLSHH211 pKa = 4.89VHH213 pKa = 6.26HH214 pKa = 6.86IKK216 pKa = 10.84YY217 pKa = 9.23MVMLQLVRR225 pKa = 11.84SVPVARR231 pKa = 11.84ALSTGDD237 pKa = 3.23AWDD240 pKa = 3.97LSLVFARR247 pKa = 11.84FLEE250 pKa = 4.47WEE252 pKa = 4.34RR253 pKa = 11.84RR254 pKa = 11.84WSLSLLDD261 pKa = 5.99LIAPYY266 pKa = 11.12LNFAWRR272 pKa = 11.84DD273 pKa = 3.74CVSPTLVKK281 pKa = 10.08TDD283 pKa = 3.2VSLGMSCSKK292 pKa = 9.64GTLVTFPSTLSLEE305 pKa = 4.72FSQGLKK311 pKa = 10.67FNVFEE316 pKa = 4.51PSEE319 pKa = 4.23LCCEE323 pKa = 4.33EE324 pKa = 3.93RR325 pKa = 11.84HH326 pKa = 6.23HH327 pKa = 6.9PLLTVSMCLAYY338 pKa = 10.73VLLWYY343 pKa = 10.21LIIQLVIISADD354 pKa = 3.52VVLDD358 pKa = 3.61ASKK361 pKa = 10.68YY362 pKa = 8.09ICEE365 pKa = 4.37TSLHH369 pKa = 5.06VCQRR373 pKa = 11.84FATLLSSLLRR383 pKa = 11.84WYY385 pKa = 10.09RR386 pKa = 11.84LKK388 pKa = 10.22IDD390 pKa = 3.5RR391 pKa = 11.84SLVRR395 pKa = 11.84DD396 pKa = 3.54SRR398 pKa = 11.84VVTSCSGEE406 pKa = 3.89APPVYY411 pKa = 10.05RR412 pKa = 11.84LVSKK416 pKa = 10.85GPEE419 pKa = 3.93AVDD422 pKa = 3.48SLEE425 pKa = 3.57MSMRR429 pKa = 11.84GSEE432 pKa = 3.66IRR434 pKa = 11.84TSVAIPGCLYY444 pKa = 10.57LAVVHH449 pKa = 7.0DD450 pKa = 4.47SDD452 pKa = 5.2VIFIGMAFRR461 pKa = 11.84YY462 pKa = 9.44KK463 pKa = 10.64GYY465 pKa = 10.62LVTAQHH471 pKa = 6.81NILAMSSAPGKK482 pKa = 10.79YY483 pKa = 8.06YY484 pKa = 10.7VLPFRR489 pKa = 11.84PGKK492 pKa = 10.27DD493 pKa = 3.2SEE495 pKa = 4.34FCYY498 pKa = 10.6LDD500 pKa = 3.31QEE502 pKa = 4.54RR503 pKa = 11.84MIEE506 pKa = 4.08LTSVMLHH513 pKa = 5.97NDD515 pKa = 2.68IVSEE519 pKa = 4.09EE520 pKa = 3.92FQGYY524 pKa = 9.47DD525 pKa = 3.18VAIIPIAAAAWSCLGVKK542 pKa = 10.12SLLHH546 pKa = 7.26ADD548 pKa = 3.68ATWGINVTLYY558 pKa = 11.14GLEE561 pKa = 3.97KK562 pKa = 9.26TGKK565 pKa = 9.88RR566 pKa = 11.84KK567 pKa = 9.0LQRR570 pKa = 11.84SLGTIRR576 pKa = 11.84EE577 pKa = 4.33DD578 pKa = 3.89KK579 pKa = 10.49EE580 pKa = 4.6SPLHH584 pKa = 4.99SVYY587 pKa = 11.19YY588 pKa = 10.02NASTLRR594 pKa = 11.84GWSGSPVLNGHH605 pKa = 5.9KK606 pKa = 9.97RR607 pKa = 11.84VVALHH612 pKa = 6.78CGTNGQVNRR621 pKa = 11.84GLNFAFVRR629 pKa = 11.84FIIEE633 pKa = 3.37IHH635 pKa = 6.5EE636 pKa = 4.34INTRR640 pKa = 11.84EE641 pKa = 4.07SSTDD645 pKa = 3.08SYY647 pKa = 11.37EE648 pKa = 4.57DD649 pKa = 3.97EE650 pKa = 4.19IEE652 pKa = 3.97KK653 pKa = 10.72SFRR656 pKa = 11.84TRR658 pKa = 11.84GGYY661 pKa = 9.8LSRR664 pKa = 11.84RR665 pKa = 11.84QEE667 pKa = 3.89EE668 pKa = 4.61LEE670 pKa = 3.86VEE672 pKa = 4.13RR673 pKa = 11.84RR674 pKa = 11.84VDD676 pKa = 3.25EE677 pKa = 5.08GIAVYY682 pKa = 10.19KK683 pKa = 10.28RR684 pKa = 11.84YY685 pKa = 9.59IVSQSSRR692 pKa = 11.84GIISFCDD699 pKa = 2.88SDD701 pKa = 4.05YY702 pKa = 11.39VDD704 pKa = 3.67RR705 pKa = 11.84QRR707 pKa = 11.84ATHH710 pKa = 6.05TATTNVVEE718 pKa = 5.37QVFDD722 pKa = 4.61DD723 pKa = 4.05NLEE726 pKa = 4.01SSYY729 pKa = 11.35SPKK732 pKa = 10.41CIQRR736 pKa = 11.84CRR738 pKa = 11.84TGEE741 pKa = 3.98AEE743 pKa = 4.04KK744 pKa = 11.2SEE746 pKa = 4.49DD747 pKa = 3.68LSSAEE752 pKa = 4.26QLFDD756 pKa = 3.64YY757 pKa = 10.88HH758 pKa = 8.62DD759 pKa = 4.58GDD761 pKa = 4.01PFVASRR767 pKa = 11.84VTTRR771 pKa = 11.84LSAPHH776 pKa = 5.56YY777 pKa = 9.01KK778 pKa = 10.17GRR780 pKa = 11.84PEE782 pKa = 4.59RR783 pKa = 11.84YY784 pKa = 9.23DD785 pKa = 3.21EE786 pKa = 4.94HH787 pKa = 8.47VDD789 pKa = 3.59LSRR792 pKa = 11.84AKK794 pKa = 10.26EE795 pKa = 3.79LGYY798 pKa = 10.86DD799 pKa = 3.54PEE801 pKa = 4.54EE802 pKa = 4.96YY803 pKa = 11.04GMPRR807 pKa = 11.84ATDD810 pKa = 3.17RR811 pKa = 11.84TSAIKK816 pKa = 10.34RR817 pKa = 11.84SAASLSKK824 pKa = 10.72SLEE827 pKa = 3.88DD828 pKa = 5.16AIRR831 pKa = 11.84TSKK834 pKa = 9.62FQKK837 pKa = 9.77PSEE840 pKa = 4.36DD841 pKa = 3.32LRR843 pKa = 11.84SAAVSIMMDD852 pKa = 3.4TLSSLRR858 pKa = 11.84YY859 pKa = 8.36PVDD862 pKa = 3.31GSGVTRR868 pKa = 11.84DD869 pKa = 4.23KK870 pKa = 10.89IASQLNSSAVGDD882 pKa = 3.91GRR884 pKa = 11.84SPGLPYY890 pKa = 10.68ASEE893 pKa = 4.36FPTNRR898 pKa = 11.84DD899 pKa = 3.36LLSKK903 pKa = 10.51FSVDD907 pKa = 3.16EE908 pKa = 4.26LADD911 pKa = 4.68LIYY914 pKa = 10.58TKK916 pKa = 10.62YY917 pKa = 10.53RR918 pKa = 11.84EE919 pKa = 4.51DD920 pKa = 3.11RR921 pKa = 11.84WEE923 pKa = 3.89QPSINFLKK931 pKa = 10.42IEE933 pKa = 3.97PTKK936 pKa = 10.78VEE938 pKa = 4.59KK939 pKa = 10.65LDD941 pKa = 3.58QGLDD945 pKa = 3.08RR946 pKa = 11.84CVQAVGLDD954 pKa = 3.31TQLYY958 pKa = 7.66FRR960 pKa = 11.84CHH962 pKa = 6.13FGALADD968 pKa = 3.84VASANYY974 pKa = 9.43RR975 pKa = 11.84KK976 pKa = 10.15SPVMQGWSPLKK987 pKa = 10.61PGDD990 pKa = 3.24GHH992 pKa = 6.15YY993 pKa = 10.24LYY995 pKa = 11.18NVITRR1000 pKa = 11.84NSNRR1004 pKa = 11.84KK1005 pKa = 8.52ILEE1008 pKa = 3.81YY1009 pKa = 10.38DD1010 pKa = 3.64GKK1012 pKa = 10.21AFEE1015 pKa = 4.78YY1016 pKa = 10.31VAHH1019 pKa = 6.03TAEE1022 pKa = 4.86AYY1024 pKa = 10.85SDD1026 pKa = 3.73VTDD1029 pKa = 4.23VIIGLSTPAVGANVDD1044 pKa = 3.91KK1045 pKa = 10.88VRR1047 pKa = 11.84EE1048 pKa = 4.2WRR1050 pKa = 11.84SEE1052 pKa = 3.59AQRR1055 pKa = 11.84FMMRR1059 pKa = 11.84VGEE1062 pKa = 4.18SGYY1065 pKa = 11.24LLADD1069 pKa = 3.39GTLVSKK1075 pKa = 10.8RR1076 pKa = 11.84SPFVLSSGRR1085 pKa = 11.84WDD1087 pKa = 3.34TFLRR1091 pKa = 11.84NSLTGYY1097 pKa = 8.16YY1098 pKa = 9.32WLIIGLLDD1106 pKa = 4.29AGYY1109 pKa = 8.15TADD1112 pKa = 5.05DD1113 pKa = 3.73IKK1115 pKa = 11.48AKK1117 pKa = 10.46FLIKK1121 pKa = 10.72VGGDD1125 pKa = 3.56DD1126 pKa = 4.53VILSVPSNFDD1136 pKa = 3.22TQTFTSALGRR1146 pKa = 11.84YY1147 pKa = 7.66GMKK1150 pKa = 9.2IHH1152 pKa = 6.72KK1153 pKa = 9.99VKK1155 pKa = 10.76LKK1157 pKa = 10.52PITDD1161 pKa = 3.67DD1162 pKa = 4.42FEE1164 pKa = 4.42FFSWKK1169 pKa = 9.68FSKK1172 pKa = 10.62NSRR1175 pKa = 11.84GLVQWTPTRR1184 pKa = 11.84FSKK1187 pKa = 10.81HH1188 pKa = 5.81LEE1190 pKa = 3.74NFFNTKK1196 pKa = 10.24HH1197 pKa = 6.38EE1198 pKa = 4.65DD1199 pKa = 2.9RR1200 pKa = 11.84HH1201 pKa = 5.2QALVSHH1207 pKa = 6.22MMNWVHH1213 pKa = 6.91SEE1215 pKa = 3.27PHH1217 pKa = 5.82YY1218 pKa = 11.09NFFRR1222 pKa = 11.84EE1223 pKa = 3.65IYY1225 pKa = 8.02MRR1227 pKa = 11.84GRR1229 pKa = 11.84EE1230 pKa = 4.32SEE1232 pKa = 3.88PDD1234 pKa = 2.96IYY1236 pKa = 10.6EE1237 pKa = 3.8LSKK1240 pKa = 10.63IPEE1243 pKa = 3.97RR1244 pKa = 11.84RR1245 pKa = 11.84DD1246 pKa = 3.02LLFYY1250 pKa = 11.08LRR1252 pKa = 11.84GNEE1255 pKa = 4.6SVIKK1259 pKa = 10.41DD1260 pKa = 3.55WGSVGAFVSLASAAVLKK1277 pKa = 11.14NNN1279 pKa = 4.11
MM1 pKa = 7.42EE2 pKa = 6.21PKK4 pKa = 9.27TQVNYY9 pKa = 11.13AEE11 pKa = 4.53VVGGRR16 pKa = 11.84YY17 pKa = 8.67NRR19 pKa = 11.84PAEE22 pKa = 3.99VRR24 pKa = 11.84AMRR27 pKa = 11.84FLASNLYY34 pKa = 9.88GAGIVFDD41 pKa = 3.96ATEE44 pKa = 4.43GGAKK48 pKa = 10.09CFTSPPPHH56 pKa = 6.66PASLRR61 pKa = 11.84FCSVRR66 pKa = 11.84GMPALAVCQTQGWWASFVLKK86 pKa = 10.51SLPFPARR93 pKa = 11.84WLIFQFLNYY102 pKa = 8.76CWEE105 pKa = 4.0WVEE108 pKa = 3.96LQDD111 pKa = 4.46RR112 pKa = 11.84SAVFTATTWTLLPMAKK128 pKa = 9.43RR129 pKa = 11.84FMGPGLEE136 pKa = 4.09YY137 pKa = 10.61PEE139 pKa = 5.73IGEE142 pKa = 4.29DD143 pKa = 2.92WFDD146 pKa = 5.55RR147 pKa = 11.84IIDD150 pKa = 3.74SPRR153 pKa = 11.84YY154 pKa = 9.28RR155 pKa = 11.84GWEE158 pKa = 3.81VVPQVSYY165 pKa = 10.9LYY167 pKa = 10.68QNVGNCCLSYY177 pKa = 10.42SAHH180 pKa = 5.82EE181 pKa = 4.6KK182 pKa = 9.81IVAFVTEE189 pKa = 4.95LYY191 pKa = 10.75CRR193 pKa = 11.84FSTYY197 pKa = 10.78GIYY200 pKa = 10.67LSARR204 pKa = 11.84TLRR207 pKa = 11.84QLSHH211 pKa = 4.89VHH213 pKa = 6.26HH214 pKa = 6.86IKK216 pKa = 10.84YY217 pKa = 9.23MVMLQLVRR225 pKa = 11.84SVPVARR231 pKa = 11.84ALSTGDD237 pKa = 3.23AWDD240 pKa = 3.97LSLVFARR247 pKa = 11.84FLEE250 pKa = 4.47WEE252 pKa = 4.34RR253 pKa = 11.84RR254 pKa = 11.84WSLSLLDD261 pKa = 5.99LIAPYY266 pKa = 11.12LNFAWRR272 pKa = 11.84DD273 pKa = 3.74CVSPTLVKK281 pKa = 10.08TDD283 pKa = 3.2VSLGMSCSKK292 pKa = 9.64GTLVTFPSTLSLEE305 pKa = 4.72FSQGLKK311 pKa = 10.67FNVFEE316 pKa = 4.51PSEE319 pKa = 4.23LCCEE323 pKa = 4.33EE324 pKa = 3.93RR325 pKa = 11.84HH326 pKa = 6.23HH327 pKa = 6.9PLLTVSMCLAYY338 pKa = 10.73VLLWYY343 pKa = 10.21LIIQLVIISADD354 pKa = 3.52VVLDD358 pKa = 3.61ASKK361 pKa = 10.68YY362 pKa = 8.09ICEE365 pKa = 4.37TSLHH369 pKa = 5.06VCQRR373 pKa = 11.84FATLLSSLLRR383 pKa = 11.84WYY385 pKa = 10.09RR386 pKa = 11.84LKK388 pKa = 10.22IDD390 pKa = 3.5RR391 pKa = 11.84SLVRR395 pKa = 11.84DD396 pKa = 3.54SRR398 pKa = 11.84VVTSCSGEE406 pKa = 3.89APPVYY411 pKa = 10.05RR412 pKa = 11.84LVSKK416 pKa = 10.85GPEE419 pKa = 3.93AVDD422 pKa = 3.48SLEE425 pKa = 3.57MSMRR429 pKa = 11.84GSEE432 pKa = 3.66IRR434 pKa = 11.84TSVAIPGCLYY444 pKa = 10.57LAVVHH449 pKa = 7.0DD450 pKa = 4.47SDD452 pKa = 5.2VIFIGMAFRR461 pKa = 11.84YY462 pKa = 9.44KK463 pKa = 10.64GYY465 pKa = 10.62LVTAQHH471 pKa = 6.81NILAMSSAPGKK482 pKa = 10.79YY483 pKa = 8.06YY484 pKa = 10.7VLPFRR489 pKa = 11.84PGKK492 pKa = 10.27DD493 pKa = 3.2SEE495 pKa = 4.34FCYY498 pKa = 10.6LDD500 pKa = 3.31QEE502 pKa = 4.54RR503 pKa = 11.84MIEE506 pKa = 4.08LTSVMLHH513 pKa = 5.97NDD515 pKa = 2.68IVSEE519 pKa = 4.09EE520 pKa = 3.92FQGYY524 pKa = 9.47DD525 pKa = 3.18VAIIPIAAAAWSCLGVKK542 pKa = 10.12SLLHH546 pKa = 7.26ADD548 pKa = 3.68ATWGINVTLYY558 pKa = 11.14GLEE561 pKa = 3.97KK562 pKa = 9.26TGKK565 pKa = 9.88RR566 pKa = 11.84KK567 pKa = 9.0LQRR570 pKa = 11.84SLGTIRR576 pKa = 11.84EE577 pKa = 4.33DD578 pKa = 3.89KK579 pKa = 10.49EE580 pKa = 4.6SPLHH584 pKa = 4.99SVYY587 pKa = 11.19YY588 pKa = 10.02NASTLRR594 pKa = 11.84GWSGSPVLNGHH605 pKa = 5.9KK606 pKa = 9.97RR607 pKa = 11.84VVALHH612 pKa = 6.78CGTNGQVNRR621 pKa = 11.84GLNFAFVRR629 pKa = 11.84FIIEE633 pKa = 3.37IHH635 pKa = 6.5EE636 pKa = 4.34INTRR640 pKa = 11.84EE641 pKa = 4.07SSTDD645 pKa = 3.08SYY647 pKa = 11.37EE648 pKa = 4.57DD649 pKa = 3.97EE650 pKa = 4.19IEE652 pKa = 3.97KK653 pKa = 10.72SFRR656 pKa = 11.84TRR658 pKa = 11.84GGYY661 pKa = 9.8LSRR664 pKa = 11.84RR665 pKa = 11.84QEE667 pKa = 3.89EE668 pKa = 4.61LEE670 pKa = 3.86VEE672 pKa = 4.13RR673 pKa = 11.84RR674 pKa = 11.84VDD676 pKa = 3.25EE677 pKa = 5.08GIAVYY682 pKa = 10.19KK683 pKa = 10.28RR684 pKa = 11.84YY685 pKa = 9.59IVSQSSRR692 pKa = 11.84GIISFCDD699 pKa = 2.88SDD701 pKa = 4.05YY702 pKa = 11.39VDD704 pKa = 3.67RR705 pKa = 11.84QRR707 pKa = 11.84ATHH710 pKa = 6.05TATTNVVEE718 pKa = 5.37QVFDD722 pKa = 4.61DD723 pKa = 4.05NLEE726 pKa = 4.01SSYY729 pKa = 11.35SPKK732 pKa = 10.41CIQRR736 pKa = 11.84CRR738 pKa = 11.84TGEE741 pKa = 3.98AEE743 pKa = 4.04KK744 pKa = 11.2SEE746 pKa = 4.49DD747 pKa = 3.68LSSAEE752 pKa = 4.26QLFDD756 pKa = 3.64YY757 pKa = 10.88HH758 pKa = 8.62DD759 pKa = 4.58GDD761 pKa = 4.01PFVASRR767 pKa = 11.84VTTRR771 pKa = 11.84LSAPHH776 pKa = 5.56YY777 pKa = 9.01KK778 pKa = 10.17GRR780 pKa = 11.84PEE782 pKa = 4.59RR783 pKa = 11.84YY784 pKa = 9.23DD785 pKa = 3.21EE786 pKa = 4.94HH787 pKa = 8.47VDD789 pKa = 3.59LSRR792 pKa = 11.84AKK794 pKa = 10.26EE795 pKa = 3.79LGYY798 pKa = 10.86DD799 pKa = 3.54PEE801 pKa = 4.54EE802 pKa = 4.96YY803 pKa = 11.04GMPRR807 pKa = 11.84ATDD810 pKa = 3.17RR811 pKa = 11.84TSAIKK816 pKa = 10.34RR817 pKa = 11.84SAASLSKK824 pKa = 10.72SLEE827 pKa = 3.88DD828 pKa = 5.16AIRR831 pKa = 11.84TSKK834 pKa = 9.62FQKK837 pKa = 9.77PSEE840 pKa = 4.36DD841 pKa = 3.32LRR843 pKa = 11.84SAAVSIMMDD852 pKa = 3.4TLSSLRR858 pKa = 11.84YY859 pKa = 8.36PVDD862 pKa = 3.31GSGVTRR868 pKa = 11.84DD869 pKa = 4.23KK870 pKa = 10.89IASQLNSSAVGDD882 pKa = 3.91GRR884 pKa = 11.84SPGLPYY890 pKa = 10.68ASEE893 pKa = 4.36FPTNRR898 pKa = 11.84DD899 pKa = 3.36LLSKK903 pKa = 10.51FSVDD907 pKa = 3.16EE908 pKa = 4.26LADD911 pKa = 4.68LIYY914 pKa = 10.58TKK916 pKa = 10.62YY917 pKa = 10.53RR918 pKa = 11.84EE919 pKa = 4.51DD920 pKa = 3.11RR921 pKa = 11.84WEE923 pKa = 3.89QPSINFLKK931 pKa = 10.42IEE933 pKa = 3.97PTKK936 pKa = 10.78VEE938 pKa = 4.59KK939 pKa = 10.65LDD941 pKa = 3.58QGLDD945 pKa = 3.08RR946 pKa = 11.84CVQAVGLDD954 pKa = 3.31TQLYY958 pKa = 7.66FRR960 pKa = 11.84CHH962 pKa = 6.13FGALADD968 pKa = 3.84VASANYY974 pKa = 9.43RR975 pKa = 11.84KK976 pKa = 10.15SPVMQGWSPLKK987 pKa = 10.61PGDD990 pKa = 3.24GHH992 pKa = 6.15YY993 pKa = 10.24LYY995 pKa = 11.18NVITRR1000 pKa = 11.84NSNRR1004 pKa = 11.84KK1005 pKa = 8.52ILEE1008 pKa = 3.81YY1009 pKa = 10.38DD1010 pKa = 3.64GKK1012 pKa = 10.21AFEE1015 pKa = 4.78YY1016 pKa = 10.31VAHH1019 pKa = 6.03TAEE1022 pKa = 4.86AYY1024 pKa = 10.85SDD1026 pKa = 3.73VTDD1029 pKa = 4.23VIIGLSTPAVGANVDD1044 pKa = 3.91KK1045 pKa = 10.88VRR1047 pKa = 11.84EE1048 pKa = 4.2WRR1050 pKa = 11.84SEE1052 pKa = 3.59AQRR1055 pKa = 11.84FMMRR1059 pKa = 11.84VGEE1062 pKa = 4.18SGYY1065 pKa = 11.24LLADD1069 pKa = 3.39GTLVSKK1075 pKa = 10.8RR1076 pKa = 11.84SPFVLSSGRR1085 pKa = 11.84WDD1087 pKa = 3.34TFLRR1091 pKa = 11.84NSLTGYY1097 pKa = 8.16YY1098 pKa = 9.32WLIIGLLDD1106 pKa = 4.29AGYY1109 pKa = 8.15TADD1112 pKa = 5.05DD1113 pKa = 3.73IKK1115 pKa = 11.48AKK1117 pKa = 10.46FLIKK1121 pKa = 10.72VGGDD1125 pKa = 3.56DD1126 pKa = 4.53VILSVPSNFDD1136 pKa = 3.22TQTFTSALGRR1146 pKa = 11.84YY1147 pKa = 7.66GMKK1150 pKa = 9.2IHH1152 pKa = 6.72KK1153 pKa = 9.99VKK1155 pKa = 10.76LKK1157 pKa = 10.52PITDD1161 pKa = 3.67DD1162 pKa = 4.42FEE1164 pKa = 4.42FFSWKK1169 pKa = 9.68FSKK1172 pKa = 10.62NSRR1175 pKa = 11.84GLVQWTPTRR1184 pKa = 11.84FSKK1187 pKa = 10.81HH1188 pKa = 5.81LEE1190 pKa = 3.74NFFNTKK1196 pKa = 10.24HH1197 pKa = 6.38EE1198 pKa = 4.65DD1199 pKa = 2.9RR1200 pKa = 11.84HH1201 pKa = 5.2QALVSHH1207 pKa = 6.22MMNWVHH1213 pKa = 6.91SEE1215 pKa = 3.27PHH1217 pKa = 5.82YY1218 pKa = 11.09NFFRR1222 pKa = 11.84EE1223 pKa = 3.65IYY1225 pKa = 8.02MRR1227 pKa = 11.84GRR1229 pKa = 11.84EE1230 pKa = 4.32SEE1232 pKa = 3.88PDD1234 pKa = 2.96IYY1236 pKa = 10.6EE1237 pKa = 3.8LSKK1240 pKa = 10.63IPEE1243 pKa = 3.97RR1244 pKa = 11.84RR1245 pKa = 11.84DD1246 pKa = 3.02LLFYY1250 pKa = 11.08LRR1252 pKa = 11.84GNEE1255 pKa = 4.6SVIKK1259 pKa = 10.41DD1260 pKa = 3.55WGSVGAFVSLASAAVLKK1277 pKa = 11.14NNN1279 pKa = 4.11
Molecular weight: 145.09 kDa
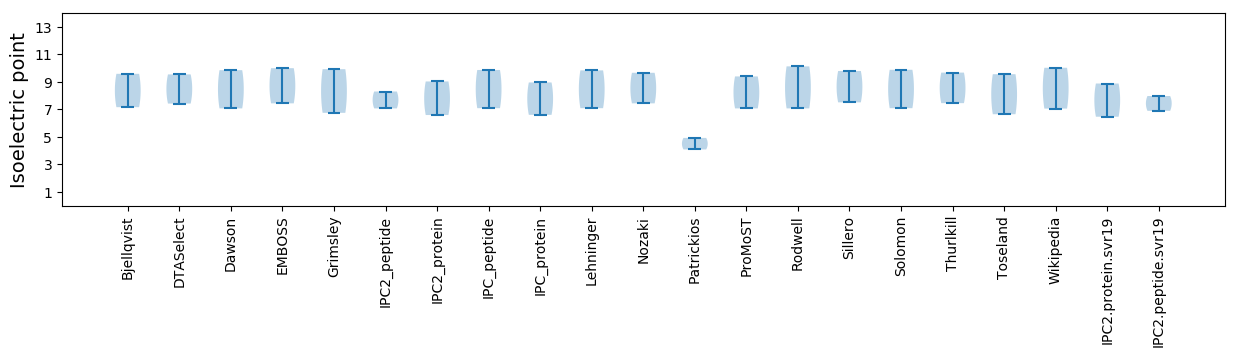
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9WTA1|Q9WTA1_9VIRU Polyprotein OS=Sclerophthora macrospora virus B OX=75914 PE=4 SV=1
MM1 pKa = 7.14TKK3 pKa = 10.66KK4 pKa = 10.21NVKK7 pKa = 8.41TSGAIVVYY15 pKa = 10.23KK16 pKa = 9.88PQTRR20 pKa = 11.84LPPQNKK26 pKa = 9.19KK27 pKa = 10.58KK28 pKa = 10.25NRR30 pKa = 11.84KK31 pKa = 8.29KK32 pKa = 10.36KK33 pKa = 10.3GRR35 pKa = 11.84VGPGKK40 pKa = 10.53SDD42 pKa = 3.14MLACYY47 pKa = 9.26RR48 pKa = 11.84ASLSAPFSITAQGARR63 pKa = 11.84VPDD66 pKa = 4.29MYY68 pKa = 11.21SCPTVTRR75 pKa = 11.84HH76 pKa = 4.26ITKK79 pKa = 10.36SFTVVTNSAGEE90 pKa = 3.89ADD92 pKa = 4.47LVVLPSAFHH101 pKa = 6.32HH102 pKa = 6.03AVSPRR107 pKa = 11.84NNIPSGVTWSTLSGATAANALVFTSPGQLANSLVNYY143 pKa = 9.65RR144 pKa = 11.84IVGYY148 pKa = 7.74GVRR151 pKa = 11.84IFGVASMTNTAGRR164 pKa = 11.84AVIATVPIASWINDD178 pKa = 3.03KK179 pKa = 9.49TASVGGQVSNAVNAAANVAGTLVAYY204 pKa = 7.58GVPVTGAYY212 pKa = 10.18VDD214 pKa = 3.81IASLPSLPNTVGVSMMNLSEE234 pKa = 4.24RR235 pKa = 11.84QMIITPKK242 pKa = 8.87ITGPEE247 pKa = 3.37AFVFNEE253 pKa = 4.19TTDD256 pKa = 3.34NAIGFNIVDD265 pKa = 3.6QTSVSFVASGDD276 pKa = 3.37ASYY279 pKa = 11.38LRR281 pKa = 11.84VAGFEE286 pKa = 4.16GVVIGLSGCPPNVGVLEE303 pKa = 4.3VEE305 pKa = 4.36LVYY308 pKa = 10.73HH309 pKa = 6.93LEE311 pKa = 4.26GQPTITNNNIVGDD324 pKa = 4.17SPEE327 pKa = 4.31SFCAPVSFLNTLQAIAKK344 pKa = 7.77TPTFKK349 pKa = 10.59QGLKK353 pKa = 10.25VVGNSIYY360 pKa = 10.39PGLGTLASRR369 pKa = 11.84YY370 pKa = 9.18
MM1 pKa = 7.14TKK3 pKa = 10.66KK4 pKa = 10.21NVKK7 pKa = 8.41TSGAIVVYY15 pKa = 10.23KK16 pKa = 9.88PQTRR20 pKa = 11.84LPPQNKK26 pKa = 9.19KK27 pKa = 10.58KK28 pKa = 10.25NRR30 pKa = 11.84KK31 pKa = 8.29KK32 pKa = 10.36KK33 pKa = 10.3GRR35 pKa = 11.84VGPGKK40 pKa = 10.53SDD42 pKa = 3.14MLACYY47 pKa = 9.26RR48 pKa = 11.84ASLSAPFSITAQGARR63 pKa = 11.84VPDD66 pKa = 4.29MYY68 pKa = 11.21SCPTVTRR75 pKa = 11.84HH76 pKa = 4.26ITKK79 pKa = 10.36SFTVVTNSAGEE90 pKa = 3.89ADD92 pKa = 4.47LVVLPSAFHH101 pKa = 6.32HH102 pKa = 6.03AVSPRR107 pKa = 11.84NNIPSGVTWSTLSGATAANALVFTSPGQLANSLVNYY143 pKa = 9.65RR144 pKa = 11.84IVGYY148 pKa = 7.74GVRR151 pKa = 11.84IFGVASMTNTAGRR164 pKa = 11.84AVIATVPIASWINDD178 pKa = 3.03KK179 pKa = 9.49TASVGGQVSNAVNAAANVAGTLVAYY204 pKa = 7.58GVPVTGAYY212 pKa = 10.18VDD214 pKa = 3.81IASLPSLPNTVGVSMMNLSEE234 pKa = 4.24RR235 pKa = 11.84QMIITPKK242 pKa = 8.87ITGPEE247 pKa = 3.37AFVFNEE253 pKa = 4.19TTDD256 pKa = 3.34NAIGFNIVDD265 pKa = 3.6QTSVSFVASGDD276 pKa = 3.37ASYY279 pKa = 11.38LRR281 pKa = 11.84VAGFEE286 pKa = 4.16GVVIGLSGCPPNVGVLEE303 pKa = 4.3VEE305 pKa = 4.36LVYY308 pKa = 10.73HH309 pKa = 6.93LEE311 pKa = 4.26GQPTITNNNIVGDD324 pKa = 4.17SPEE327 pKa = 4.31SFCAPVSFLNTLQAIAKK344 pKa = 7.77TPTFKK349 pKa = 10.59QGLKK353 pKa = 10.25VVGNSIYY360 pKa = 10.39PGLGTLASRR369 pKa = 11.84YY370 pKa = 9.18
Molecular weight: 38.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1649 |
370 |
1279 |
824.5 |
91.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.58 ± 1.376 | 1.698 ± 0.316 |
4.912 ± 1.269 | 5.276 ± 1.455 |
4.306 ± 0.405 | 6.671 ± 1.15 |
2.062 ± 0.502 | 4.791 ± 0.315 |
4.488 ± 0.055 | 8.914 ± 1.381 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.941 ± 0.025 | 3.76 ± 1.534 |
4.73 ± 0.899 | 2.668 ± 0.018 |
6.367 ± 1.46 | 9.46 ± 0.277 |
5.761 ± 1.339 | 8.611 ± 1.679 |
1.698 ± 0.592 | 4.306 ± 0.682 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
