
gamma proteobacterium SS-5
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; unclassified Gammaproteobacteria
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
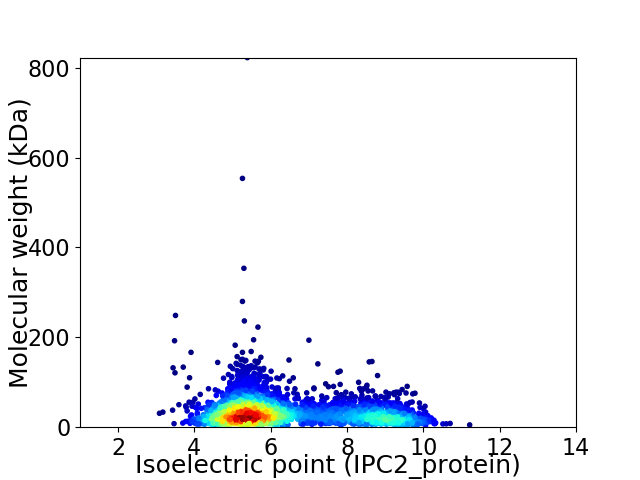
Virtual 2D-PAGE plot for 3199 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
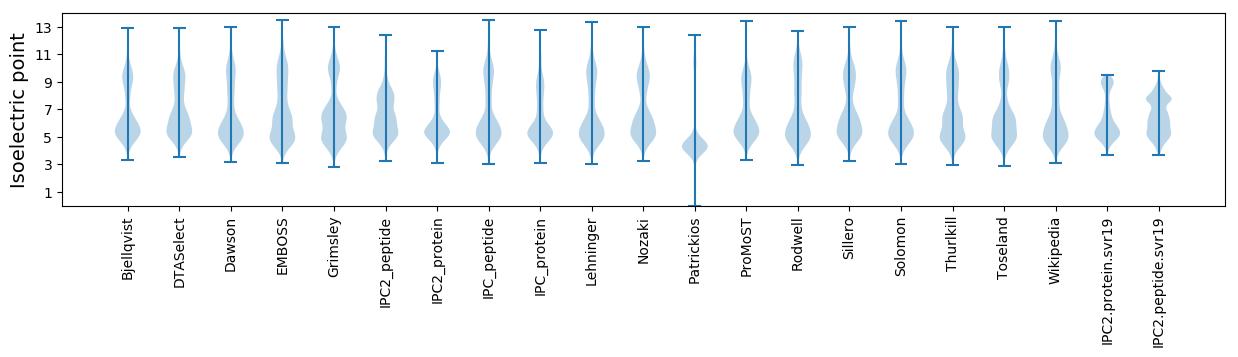
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5Q0ELG3|A0A5Q0ELG3_9GAMM Ribosomal RNA small subunit methyltransferase D OS=gamma proteobacterium SS-5 OX=947516 GN=rsmD PE=3 SV=1
MM1 pKa = 7.11SHH3 pKa = 6.88HH4 pKa = 6.86LAAEE8 pKa = 4.12LVFTYY13 pKa = 10.66QEE15 pKa = 4.03VLDD18 pKa = 4.31ALQAGFGQLSNTPVGTSFSTVEE40 pKa = 3.66LLANGFSITLDD51 pKa = 3.56GSFVWNGTVPEE62 pKa = 4.66GSLKK66 pKa = 10.59RR67 pKa = 11.84LFVSHH72 pKa = 6.44EE73 pKa = 3.76RR74 pKa = 11.84DD75 pKa = 3.12AAALEE80 pKa = 4.3GQISISDD87 pKa = 3.85SATLLLTGIASSFAWGDD104 pKa = 3.32YY105 pKa = 10.91DD106 pKa = 3.75ATTTEE111 pKa = 4.14FSNVKK116 pKa = 8.55TVSGFSLDD124 pKa = 3.58MAQVASIGFEE134 pKa = 4.13LLMEE138 pKa = 5.22ANTAVTATDD147 pKa = 3.78GDD149 pKa = 4.09DD150 pKa = 3.55TFDD153 pKa = 3.46GSGAADD159 pKa = 3.38VVYY162 pKa = 10.66SGAGSDD168 pKa = 3.32ILRR171 pKa = 11.84GGDD174 pKa = 3.88GNDD177 pKa = 3.0ILYY180 pKa = 10.97GEE182 pKa = 4.99AGNDD186 pKa = 3.27FLYY189 pKa = 10.65GGRR192 pKa = 11.84GNDD195 pKa = 3.27EE196 pKa = 4.73LYY198 pKa = 10.91GGTGADD204 pKa = 3.68YY205 pKa = 10.88MEE207 pKa = 5.42GGSGADD213 pKa = 3.8KK214 pKa = 11.27YY215 pKa = 11.2FIDD218 pKa = 6.09DD219 pKa = 3.46IGDD222 pKa = 3.68QVVEE226 pKa = 4.04TDD228 pKa = 3.86NNPEE232 pKa = 3.95EE233 pKa = 4.54PEE235 pKa = 3.66AAARR239 pKa = 11.84ILLALDD245 pKa = 3.43ISSTIDD251 pKa = 3.1QVFASIDD258 pKa = 3.68YY259 pKa = 11.02ALTAYY264 pKa = 10.43VEE266 pKa = 4.15NLTLTDD272 pKa = 3.39NAVQATGNEE281 pKa = 4.13LDD283 pKa = 4.45NILTGSSANNILTGGAGNDD302 pKa = 4.27TIDD305 pKa = 3.65GGAGVDD311 pKa = 3.02IAVFSGNHH319 pKa = 5.3SSYY322 pKa = 10.87SINGSAGSYY331 pKa = 9.45TLTGPDD337 pKa = 3.46GTDD340 pKa = 2.51TLTNIEE346 pKa = 4.32RR347 pKa = 11.84LQFADD352 pKa = 4.25KK353 pKa = 10.75KK354 pKa = 10.61LAYY357 pKa = 10.18DD358 pKa = 4.63LGATEE363 pKa = 4.1NAGMSLEE370 pKa = 4.47FINVIASSLVNDD382 pKa = 4.03ASTRR386 pKa = 11.84GLILNFFDD394 pKa = 4.77QGHH397 pKa = 6.07SLNSLFQLAVDD408 pKa = 3.98IGLVSTLAGGSSNEE422 pKa = 3.92AVAQLAYY429 pKa = 10.66RR430 pKa = 11.84NLLGTEE436 pKa = 4.14ADD438 pKa = 3.59QATTATLTGYY448 pKa = 10.54VDD450 pKa = 3.54IMGQAGFLAAVAGMEE465 pKa = 4.16INQQAVGLLGLQQAGMEE482 pKa = 4.27YY483 pKa = 10.8LL484 pKa = 3.67
MM1 pKa = 7.11SHH3 pKa = 6.88HH4 pKa = 6.86LAAEE8 pKa = 4.12LVFTYY13 pKa = 10.66QEE15 pKa = 4.03VLDD18 pKa = 4.31ALQAGFGQLSNTPVGTSFSTVEE40 pKa = 3.66LLANGFSITLDD51 pKa = 3.56GSFVWNGTVPEE62 pKa = 4.66GSLKK66 pKa = 10.59RR67 pKa = 11.84LFVSHH72 pKa = 6.44EE73 pKa = 3.76RR74 pKa = 11.84DD75 pKa = 3.12AAALEE80 pKa = 4.3GQISISDD87 pKa = 3.85SATLLLTGIASSFAWGDD104 pKa = 3.32YY105 pKa = 10.91DD106 pKa = 3.75ATTTEE111 pKa = 4.14FSNVKK116 pKa = 8.55TVSGFSLDD124 pKa = 3.58MAQVASIGFEE134 pKa = 4.13LLMEE138 pKa = 5.22ANTAVTATDD147 pKa = 3.78GDD149 pKa = 4.09DD150 pKa = 3.55TFDD153 pKa = 3.46GSGAADD159 pKa = 3.38VVYY162 pKa = 10.66SGAGSDD168 pKa = 3.32ILRR171 pKa = 11.84GGDD174 pKa = 3.88GNDD177 pKa = 3.0ILYY180 pKa = 10.97GEE182 pKa = 4.99AGNDD186 pKa = 3.27FLYY189 pKa = 10.65GGRR192 pKa = 11.84GNDD195 pKa = 3.27EE196 pKa = 4.73LYY198 pKa = 10.91GGTGADD204 pKa = 3.68YY205 pKa = 10.88MEE207 pKa = 5.42GGSGADD213 pKa = 3.8KK214 pKa = 11.27YY215 pKa = 11.2FIDD218 pKa = 6.09DD219 pKa = 3.46IGDD222 pKa = 3.68QVVEE226 pKa = 4.04TDD228 pKa = 3.86NNPEE232 pKa = 3.95EE233 pKa = 4.54PEE235 pKa = 3.66AAARR239 pKa = 11.84ILLALDD245 pKa = 3.43ISSTIDD251 pKa = 3.1QVFASIDD258 pKa = 3.68YY259 pKa = 11.02ALTAYY264 pKa = 10.43VEE266 pKa = 4.15NLTLTDD272 pKa = 3.39NAVQATGNEE281 pKa = 4.13LDD283 pKa = 4.45NILTGSSANNILTGGAGNDD302 pKa = 4.27TIDD305 pKa = 3.65GGAGVDD311 pKa = 3.02IAVFSGNHH319 pKa = 5.3SSYY322 pKa = 10.87SINGSAGSYY331 pKa = 9.45TLTGPDD337 pKa = 3.46GTDD340 pKa = 2.51TLTNIEE346 pKa = 4.32RR347 pKa = 11.84LQFADD352 pKa = 4.25KK353 pKa = 10.75KK354 pKa = 10.61LAYY357 pKa = 10.18DD358 pKa = 4.63LGATEE363 pKa = 4.1NAGMSLEE370 pKa = 4.47FINVIASSLVNDD382 pKa = 4.03ASTRR386 pKa = 11.84GLILNFFDD394 pKa = 4.77QGHH397 pKa = 6.07SLNSLFQLAVDD408 pKa = 3.98IGLVSTLAGGSSNEE422 pKa = 3.92AVAQLAYY429 pKa = 10.66RR430 pKa = 11.84NLLGTEE436 pKa = 4.14ADD438 pKa = 3.59QATTATLTGYY448 pKa = 10.54VDD450 pKa = 3.54IMGQAGFLAAVAGMEE465 pKa = 4.16INQQAVGLLGLQQAGMEE482 pKa = 4.27YY483 pKa = 10.8LL484 pKa = 3.67
Molecular weight: 50.13 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q0EJS8|A0A5Q0EJS8_9GAMM Bifunctional diguanylate cyclase/phosphodiesterase OS=gamma proteobacterium SS-5 OX=947516 GN=D5125_09210 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.91GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.25NGRR28 pKa = 11.84KK29 pKa = 9.25VLSARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.85GRR39 pKa = 11.84KK40 pKa = 8.98RR41 pKa = 11.84LTVV44 pKa = 3.11
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.91GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.25NGRR28 pKa = 11.84KK29 pKa = 9.25VLSARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.85GRR39 pKa = 11.84KK40 pKa = 8.98RR41 pKa = 11.84LTVV44 pKa = 3.11
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1044158 |
37 |
7620 |
326.4 |
36.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.357 ± 0.071 | 1.035 ± 0.019 |
5.6 ± 0.033 | 6.389 ± 0.047 |
3.447 ± 0.027 | 7.827 ± 0.043 |
2.282 ± 0.02 | 5.072 ± 0.037 |
3.447 ± 0.045 | 12.256 ± 0.066 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.325 ± 0.021 | 2.898 ± 0.03 |
5.0 ± 0.042 | 5.524 ± 0.048 |
7.114 ± 0.044 | 5.505 ± 0.033 |
4.063 ± 0.044 | 6.025 ± 0.033 |
1.298 ± 0.019 | 2.534 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
