
Lysobacter sp. Root494
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Lysobacter; unclassified Lysobacter
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
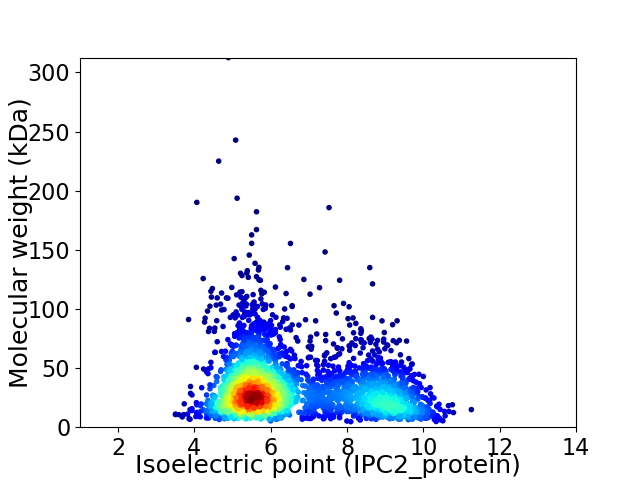
Virtual 2D-PAGE plot for 3007 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q7PEV1|A0A0Q7PEV1_9GAMM Uncharacterized protein OS=Lysobacter sp. Root494 OX=1736549 GN=ASD14_14350 PE=4 SV=1
MM1 pKa = 7.52PNAVVVNGQPSQPNQNQGGLFQWSFVGAPVGTLANANSPQVQLTPFDD48 pKa = 3.88VAASTQGILRR58 pKa = 11.84LTVTVSGCPGISTKK72 pKa = 10.6DD73 pKa = 3.4VPVTVTNAHH82 pKa = 6.3DD83 pKa = 3.82VVINLPPHH91 pKa = 6.44AVPVASPSTASEE103 pKa = 4.23GMVVTLDD110 pKa = 4.38GSGSWDD116 pKa = 3.19VDD118 pKa = 3.83NPSLTYY124 pKa = 10.76AWLQTVGPAVTLANTGNPAIKK145 pKa = 9.78TFVAPNFADD154 pKa = 3.68DD155 pKa = 3.68TQLTFRR161 pKa = 11.84LTVSDD166 pKa = 3.86GTLSNSDD173 pKa = 3.3TTFVNVSWTNDD184 pKa = 3.15SPVAGLVCPDD194 pKa = 2.99GMFEE198 pKa = 4.21VNEE201 pKa = 4.33GQSVTFDD208 pKa = 3.43ASGSTDD214 pKa = 3.41SDD216 pKa = 3.78DD217 pKa = 5.86GIATYY222 pKa = 10.09EE223 pKa = 3.88WAQLVGLPEE232 pKa = 4.15VTGVADD238 pKa = 3.43WNTAVVTFNAPQLGFGEE255 pKa = 4.46TGLVPFKK262 pKa = 10.71LTVTDD267 pKa = 3.16HH268 pKa = 6.91HH269 pKa = 6.49GAKK272 pKa = 8.77STANCAIFVHH282 pKa = 7.29DD283 pKa = 3.52ITKK286 pKa = 10.0PLLSVPQSPLVAEE299 pKa = 4.36ATSADD304 pKa = 3.89GANVAYY310 pKa = 9.63EE311 pKa = 4.18VSAFDD316 pKa = 5.45AVDD319 pKa = 3.51GDD321 pKa = 4.55LPFPSPYY328 pKa = 10.26LLCEE332 pKa = 4.37PPPGSLFALDD342 pKa = 3.71SLSPVLCSAEE352 pKa = 4.04DD353 pKa = 3.41SAGNGNTATFSVSVVDD369 pKa = 3.82STAPAISVPLSFAVEE384 pKa = 3.95ATGPDD389 pKa = 4.07GAPADD394 pKa = 4.12YY395 pKa = 10.44VAKK398 pKa = 11.02SNDD401 pKa = 3.38AVDD404 pKa = 3.95GEE406 pKa = 4.32RR407 pKa = 11.84DD408 pKa = 4.0AVCMPASGSTFPLNTPGPTTTVHH431 pKa = 7.01CDD433 pKa = 2.76ATDD436 pKa = 3.08AHH438 pKa = 6.57GNEE441 pKa = 3.83ATTRR445 pKa = 11.84TFTVAVHH452 pKa = 6.53DD453 pKa = 4.17TTPPDD458 pKa = 3.08IDD460 pKa = 3.77AATVSADD467 pKa = 3.59LVAEE471 pKa = 4.2ATSPSGASVSFGLPSAHH488 pKa = 7.36DD489 pKa = 4.39LVDD492 pKa = 4.26LDD494 pKa = 4.84AVLVTCAPGSPHH506 pKa = 6.44VFPLGEE512 pKa = 4.11TVVQCDD518 pKa = 3.5AVDD521 pKa = 3.39SRR523 pKa = 11.84GNSTADD529 pKa = 3.27ATPGTSATFKK539 pKa = 9.61VTVRR543 pKa = 11.84DD544 pKa = 3.48TTAPVFGPVSNHH556 pKa = 4.03TVEE559 pKa = 4.37AQSGAGASFAYY570 pKa = 10.09ALPTATDD577 pKa = 3.49VVDD580 pKa = 3.77GDD582 pKa = 3.82RR583 pKa = 11.84TVTCTEE589 pKa = 4.02APALALPGVFPLGSTTVTCSASDD612 pKa = 3.32TRR614 pKa = 11.84GNGGSTSFQVTVVDD628 pKa = 4.15TTAPSLDD635 pKa = 3.46LPANITAEE643 pKa = 4.36ATGSTGAVVNFAVAASDD660 pKa = 4.71AVDD663 pKa = 3.92ADD665 pKa = 3.87VQLVCSSNSGDD676 pKa = 3.4TFALGTTTVQCIGTDD691 pKa = 3.5DD692 pKa = 4.37AGNSAGGNFTITVQDD707 pKa = 3.49TTGPAISAHH716 pKa = 5.08EE717 pKa = 4.28TVTAYY722 pKa = 8.96ATGNSAATVTYY733 pKa = 7.71TVPTAVDD740 pKa = 3.85LVDD743 pKa = 4.15GPVVVTCTPASGSSFNVGTTTVTCSAKK770 pKa = 10.44DD771 pKa = 3.48SRR773 pKa = 11.84NNPSNSTFAVTVTYY787 pKa = 10.99NFTGFFQPIDD797 pKa = 3.88NGTMNSVKK805 pKa = 10.18AGSAIPVKK813 pKa = 10.62FSLGGNQGLNIFDD826 pKa = 5.19PGPASGVIACSATDD840 pKa = 3.33GDD842 pKa = 4.73AIEE845 pKa = 4.39EE846 pKa = 4.37TVTAGNSSLQFDD858 pKa = 4.45PGSNQYY864 pKa = 10.4IYY866 pKa = 10.57VWKK869 pKa = 9.17TEE871 pKa = 4.02KK872 pKa = 10.2SWAGQCRR879 pKa = 11.84VLQVKK884 pKa = 10.09LKK886 pKa = 10.99GGTQRR891 pKa = 11.84SALFKK896 pKa = 10.82FKK898 pKa = 11.01
MM1 pKa = 7.52PNAVVVNGQPSQPNQNQGGLFQWSFVGAPVGTLANANSPQVQLTPFDD48 pKa = 3.88VAASTQGILRR58 pKa = 11.84LTVTVSGCPGISTKK72 pKa = 10.6DD73 pKa = 3.4VPVTVTNAHH82 pKa = 6.3DD83 pKa = 3.82VVINLPPHH91 pKa = 6.44AVPVASPSTASEE103 pKa = 4.23GMVVTLDD110 pKa = 4.38GSGSWDD116 pKa = 3.19VDD118 pKa = 3.83NPSLTYY124 pKa = 10.76AWLQTVGPAVTLANTGNPAIKK145 pKa = 9.78TFVAPNFADD154 pKa = 3.68DD155 pKa = 3.68TQLTFRR161 pKa = 11.84LTVSDD166 pKa = 3.86GTLSNSDD173 pKa = 3.3TTFVNVSWTNDD184 pKa = 3.15SPVAGLVCPDD194 pKa = 2.99GMFEE198 pKa = 4.21VNEE201 pKa = 4.33GQSVTFDD208 pKa = 3.43ASGSTDD214 pKa = 3.41SDD216 pKa = 3.78DD217 pKa = 5.86GIATYY222 pKa = 10.09EE223 pKa = 3.88WAQLVGLPEE232 pKa = 4.15VTGVADD238 pKa = 3.43WNTAVVTFNAPQLGFGEE255 pKa = 4.46TGLVPFKK262 pKa = 10.71LTVTDD267 pKa = 3.16HH268 pKa = 6.91HH269 pKa = 6.49GAKK272 pKa = 8.77STANCAIFVHH282 pKa = 7.29DD283 pKa = 3.52ITKK286 pKa = 10.0PLLSVPQSPLVAEE299 pKa = 4.36ATSADD304 pKa = 3.89GANVAYY310 pKa = 9.63EE311 pKa = 4.18VSAFDD316 pKa = 5.45AVDD319 pKa = 3.51GDD321 pKa = 4.55LPFPSPYY328 pKa = 10.26LLCEE332 pKa = 4.37PPPGSLFALDD342 pKa = 3.71SLSPVLCSAEE352 pKa = 4.04DD353 pKa = 3.41SAGNGNTATFSVSVVDD369 pKa = 3.82STAPAISVPLSFAVEE384 pKa = 3.95ATGPDD389 pKa = 4.07GAPADD394 pKa = 4.12YY395 pKa = 10.44VAKK398 pKa = 11.02SNDD401 pKa = 3.38AVDD404 pKa = 3.95GEE406 pKa = 4.32RR407 pKa = 11.84DD408 pKa = 4.0AVCMPASGSTFPLNTPGPTTTVHH431 pKa = 7.01CDD433 pKa = 2.76ATDD436 pKa = 3.08AHH438 pKa = 6.57GNEE441 pKa = 3.83ATTRR445 pKa = 11.84TFTVAVHH452 pKa = 6.53DD453 pKa = 4.17TTPPDD458 pKa = 3.08IDD460 pKa = 3.77AATVSADD467 pKa = 3.59LVAEE471 pKa = 4.2ATSPSGASVSFGLPSAHH488 pKa = 7.36DD489 pKa = 4.39LVDD492 pKa = 4.26LDD494 pKa = 4.84AVLVTCAPGSPHH506 pKa = 6.44VFPLGEE512 pKa = 4.11TVVQCDD518 pKa = 3.5AVDD521 pKa = 3.39SRR523 pKa = 11.84GNSTADD529 pKa = 3.27ATPGTSATFKK539 pKa = 9.61VTVRR543 pKa = 11.84DD544 pKa = 3.48TTAPVFGPVSNHH556 pKa = 4.03TVEE559 pKa = 4.37AQSGAGASFAYY570 pKa = 10.09ALPTATDD577 pKa = 3.49VVDD580 pKa = 3.77GDD582 pKa = 3.82RR583 pKa = 11.84TVTCTEE589 pKa = 4.02APALALPGVFPLGSTTVTCSASDD612 pKa = 3.32TRR614 pKa = 11.84GNGGSTSFQVTVVDD628 pKa = 4.15TTAPSLDD635 pKa = 3.46LPANITAEE643 pKa = 4.36ATGSTGAVVNFAVAASDD660 pKa = 4.71AVDD663 pKa = 3.92ADD665 pKa = 3.87VQLVCSSNSGDD676 pKa = 3.4TFALGTTTVQCIGTDD691 pKa = 3.5DD692 pKa = 4.37AGNSAGGNFTITVQDD707 pKa = 3.49TTGPAISAHH716 pKa = 5.08EE717 pKa = 4.28TVTAYY722 pKa = 8.96ATGNSAATVTYY733 pKa = 7.71TVPTAVDD740 pKa = 3.85LVDD743 pKa = 4.15GPVVVTCTPASGSSFNVGTTTVTCSAKK770 pKa = 10.44DD771 pKa = 3.48SRR773 pKa = 11.84NNPSNSTFAVTVTYY787 pKa = 10.99NFTGFFQPIDD797 pKa = 3.88NGTMNSVKK805 pKa = 10.18AGSAIPVKK813 pKa = 10.62FSLGGNQGLNIFDD826 pKa = 5.19PGPASGVIACSATDD840 pKa = 3.33GDD842 pKa = 4.73AIEE845 pKa = 4.39EE846 pKa = 4.37TVTAGNSSLQFDD858 pKa = 4.45PGSNQYY864 pKa = 10.4IYY866 pKa = 10.57VWKK869 pKa = 9.17TEE871 pKa = 4.02KK872 pKa = 10.2SWAGQCRR879 pKa = 11.84VLQVKK884 pKa = 10.09LKK886 pKa = 10.99GGTQRR891 pKa = 11.84SALFKK896 pKa = 10.82FKK898 pKa = 11.01
Molecular weight: 91.08 kDa
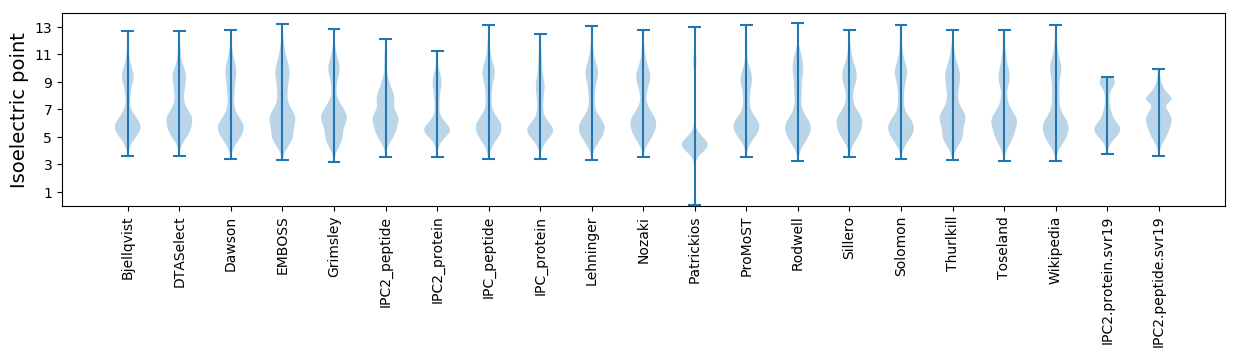
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q7Q9F8|A0A0Q7Q9F8_9GAMM Acetyl-CoA hydrolase OS=Lysobacter sp. Root494 OX=1736549 GN=ASD14_02550 PE=4 SV=1
MM1 pKa = 7.5AMKK4 pKa = 10.25KK5 pKa = 9.72AAKK8 pKa = 9.53KK9 pKa = 10.4KK10 pKa = 9.39PAAKK14 pKa = 9.97KK15 pKa = 9.35AAKK18 pKa = 9.74KK19 pKa = 9.29VAKK22 pKa = 8.89KK23 pKa = 9.1TGAKK27 pKa = 9.57KK28 pKa = 9.95AAKK31 pKa = 8.47KK32 pKa = 7.26TAKK35 pKa = 10.3KK36 pKa = 10.1AVKK39 pKa = 9.89KK40 pKa = 10.0AAKK43 pKa = 8.55KK44 pKa = 7.07TAKK47 pKa = 10.35KK48 pKa = 8.97ATKK51 pKa = 9.94KK52 pKa = 8.93VAKK55 pKa = 10.26RR56 pKa = 11.84PAKK59 pKa = 10.12KK60 pKa = 9.82AAKK63 pKa = 8.57KK64 pKa = 7.23TAKK67 pKa = 10.12KK68 pKa = 9.86AAKK71 pKa = 8.54KK72 pKa = 6.97TAKK75 pKa = 10.33RR76 pKa = 11.84PAKK79 pKa = 10.07KK80 pKa = 9.8AAKK83 pKa = 8.57KK84 pKa = 7.23TAKK87 pKa = 10.12KK88 pKa = 9.86AAKK91 pKa = 8.54KK92 pKa = 6.97TAKK95 pKa = 10.33RR96 pKa = 11.84PAKK99 pKa = 10.07KK100 pKa = 9.8AAKK103 pKa = 8.57KK104 pKa = 7.23TAKK107 pKa = 10.14KK108 pKa = 9.81AAKK111 pKa = 9.61KK112 pKa = 9.2AAKK115 pKa = 9.96RR116 pKa = 11.84PAKK119 pKa = 10.0KK120 pKa = 10.07ASKK123 pKa = 10.11KK124 pKa = 9.69RR125 pKa = 11.84ASKK128 pKa = 10.53KK129 pKa = 9.51SAAAAMPATPAPMII143 pKa = 4.39
MM1 pKa = 7.5AMKK4 pKa = 10.25KK5 pKa = 9.72AAKK8 pKa = 9.53KK9 pKa = 10.4KK10 pKa = 9.39PAAKK14 pKa = 9.97KK15 pKa = 9.35AAKK18 pKa = 9.74KK19 pKa = 9.29VAKK22 pKa = 8.89KK23 pKa = 9.1TGAKK27 pKa = 9.57KK28 pKa = 9.95AAKK31 pKa = 8.47KK32 pKa = 7.26TAKK35 pKa = 10.3KK36 pKa = 10.1AVKK39 pKa = 9.89KK40 pKa = 10.0AAKK43 pKa = 8.55KK44 pKa = 7.07TAKK47 pKa = 10.35KK48 pKa = 8.97ATKK51 pKa = 9.94KK52 pKa = 8.93VAKK55 pKa = 10.26RR56 pKa = 11.84PAKK59 pKa = 10.12KK60 pKa = 9.82AAKK63 pKa = 8.57KK64 pKa = 7.23TAKK67 pKa = 10.12KK68 pKa = 9.86AAKK71 pKa = 8.54KK72 pKa = 6.97TAKK75 pKa = 10.33RR76 pKa = 11.84PAKK79 pKa = 10.07KK80 pKa = 9.8AAKK83 pKa = 8.57KK84 pKa = 7.23TAKK87 pKa = 10.12KK88 pKa = 9.86AAKK91 pKa = 8.54KK92 pKa = 6.97TAKK95 pKa = 10.33RR96 pKa = 11.84PAKK99 pKa = 10.07KK100 pKa = 9.8AAKK103 pKa = 8.57KK104 pKa = 7.23TAKK107 pKa = 10.14KK108 pKa = 9.81AAKK111 pKa = 9.61KK112 pKa = 9.2AAKK115 pKa = 9.96RR116 pKa = 11.84PAKK119 pKa = 10.0KK120 pKa = 10.07ASKK123 pKa = 10.11KK124 pKa = 9.69RR125 pKa = 11.84ASKK128 pKa = 10.53KK129 pKa = 9.51SAAAAMPATPAPMII143 pKa = 4.39
Molecular weight: 14.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
981173 |
41 |
2982 |
326.3 |
35.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.682 ± 0.063 | 0.823 ± 0.013 |
5.875 ± 0.033 | 5.629 ± 0.044 |
3.511 ± 0.028 | 8.439 ± 0.052 |
2.29 ± 0.023 | 4.322 ± 0.028 |
3.004 ± 0.042 | 10.436 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.221 ± 0.018 | 2.685 ± 0.027 |
5.209 ± 0.031 | 3.562 ± 0.025 |
7.637 ± 0.049 | 5.263 ± 0.029 |
5.095 ± 0.039 | 7.462 ± 0.038 |
1.503 ± 0.02 | 2.352 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
