
Mulberry mosaic dwarf associated virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae
Average proteome isoelectric point is 7.33
Get precalculated fractions of proteins
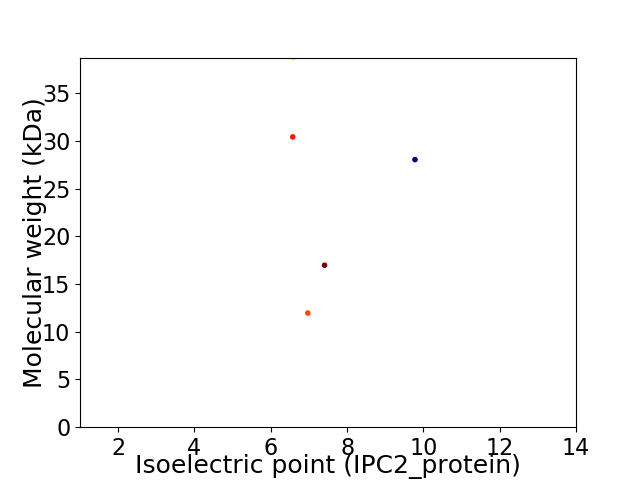
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D5BTW9|A0A0D5BTW9_9GEMI V2 protein OS=Mulberry mosaic dwarf associated virus OX=1631303 PE=4 SV=1
MM1 pKa = 7.59ASSSNFRR8 pKa = 11.84LAAKK12 pKa = 10.11NVFLTFPQCPEE23 pKa = 3.95SITWVMSHH31 pKa = 7.42LLQILNSYY39 pKa = 6.28TVKK42 pKa = 9.67YY43 pKa = 10.74ACVAEE48 pKa = 4.75EE49 pKa = 3.91KK50 pKa = 10.78HH51 pKa = 6.71KK52 pKa = 11.14DD53 pKa = 3.82DD54 pKa = 4.22NSHH57 pKa = 5.57FHH59 pKa = 7.5AIVQLDD65 pKa = 3.72KK66 pKa = 11.18KK67 pKa = 11.2LEE69 pKa = 4.05TRR71 pKa = 11.84NPNFFDD77 pKa = 4.15LPQEE81 pKa = 4.42HH82 pKa = 7.05SSGTYY87 pKa = 9.12HH88 pKa = 7.52PNVQKK93 pKa = 10.63CSSPAAVRR101 pKa = 11.84KK102 pKa = 9.1YY103 pKa = 9.62IQKK106 pKa = 8.64EE107 pKa = 3.98GHH109 pKa = 6.37FIEE112 pKa = 6.01HH113 pKa = 6.84GEE115 pKa = 3.96FNTRR119 pKa = 11.84GRR121 pKa = 11.84SPVASAEE128 pKa = 4.26KK129 pKa = 10.25IFGEE133 pKa = 4.16ILTLATDD140 pKa = 3.81EE141 pKa = 4.59EE142 pKa = 5.12SFLALVRR149 pKa = 11.84EE150 pKa = 4.55RR151 pKa = 11.84RR152 pKa = 11.84PQDD155 pKa = 3.35YY156 pKa = 10.58VLRR159 pKa = 11.84WPSITGFARR168 pKa = 11.84DD169 pKa = 3.33HH170 pKa = 5.48YY171 pKa = 10.1RR172 pKa = 11.84RR173 pKa = 11.84RR174 pKa = 11.84SIPYY178 pKa = 7.95VPRR181 pKa = 11.84WTDD184 pKa = 3.76FPGLPEE190 pKa = 6.34PIQQWAKK197 pKa = 11.29DD198 pKa = 3.78NILFEE203 pKa = 4.94PEE205 pKa = 4.33SKK207 pKa = 9.99PDD209 pKa = 3.61RR210 pKa = 11.84PISLYY215 pKa = 9.8ICGPTRR221 pKa = 11.84SGKK224 pKa = 6.56TQWARR229 pKa = 11.84SLGRR233 pKa = 11.84HH234 pKa = 5.29NYY236 pKa = 7.43FTGSLSFLDD245 pKa = 3.88YY246 pKa = 11.47DD247 pKa = 5.47DD248 pKa = 4.8FALYY252 pKa = 10.64NVIDD256 pKa = 4.96DD257 pKa = 4.16IEE259 pKa = 4.33YY260 pKa = 10.95EE261 pKa = 4.34KK262 pKa = 10.76ISTSWFKK269 pKa = 11.37SLLGAQKK276 pKa = 10.67DD277 pKa = 3.7IILKK281 pKa = 9.29GKK283 pKa = 8.39YY284 pKa = 8.83MKK286 pKa = 10.46DD287 pKa = 3.32FRR289 pKa = 11.84ISGGIPCIVLVNEE302 pKa = 4.06DD303 pKa = 3.69MDD305 pKa = 3.95WVVRR309 pKa = 11.84MASTLKK315 pKa = 10.26PWFDD319 pKa = 4.71DD320 pKa = 3.66NVLVYY325 pKa = 10.71YY326 pKa = 9.85MSASDD331 pKa = 4.0KK332 pKa = 10.91FYY334 pKa = 11.37
MM1 pKa = 7.59ASSSNFRR8 pKa = 11.84LAAKK12 pKa = 10.11NVFLTFPQCPEE23 pKa = 3.95SITWVMSHH31 pKa = 7.42LLQILNSYY39 pKa = 6.28TVKK42 pKa = 9.67YY43 pKa = 10.74ACVAEE48 pKa = 4.75EE49 pKa = 3.91KK50 pKa = 10.78HH51 pKa = 6.71KK52 pKa = 11.14DD53 pKa = 3.82DD54 pKa = 4.22NSHH57 pKa = 5.57FHH59 pKa = 7.5AIVQLDD65 pKa = 3.72KK66 pKa = 11.18KK67 pKa = 11.2LEE69 pKa = 4.05TRR71 pKa = 11.84NPNFFDD77 pKa = 4.15LPQEE81 pKa = 4.42HH82 pKa = 7.05SSGTYY87 pKa = 9.12HH88 pKa = 7.52PNVQKK93 pKa = 10.63CSSPAAVRR101 pKa = 11.84KK102 pKa = 9.1YY103 pKa = 9.62IQKK106 pKa = 8.64EE107 pKa = 3.98GHH109 pKa = 6.37FIEE112 pKa = 6.01HH113 pKa = 6.84GEE115 pKa = 3.96FNTRR119 pKa = 11.84GRR121 pKa = 11.84SPVASAEE128 pKa = 4.26KK129 pKa = 10.25IFGEE133 pKa = 4.16ILTLATDD140 pKa = 3.81EE141 pKa = 4.59EE142 pKa = 5.12SFLALVRR149 pKa = 11.84EE150 pKa = 4.55RR151 pKa = 11.84RR152 pKa = 11.84PQDD155 pKa = 3.35YY156 pKa = 10.58VLRR159 pKa = 11.84WPSITGFARR168 pKa = 11.84DD169 pKa = 3.33HH170 pKa = 5.48YY171 pKa = 10.1RR172 pKa = 11.84RR173 pKa = 11.84RR174 pKa = 11.84SIPYY178 pKa = 7.95VPRR181 pKa = 11.84WTDD184 pKa = 3.76FPGLPEE190 pKa = 6.34PIQQWAKK197 pKa = 11.29DD198 pKa = 3.78NILFEE203 pKa = 4.94PEE205 pKa = 4.33SKK207 pKa = 9.99PDD209 pKa = 3.61RR210 pKa = 11.84PISLYY215 pKa = 9.8ICGPTRR221 pKa = 11.84SGKK224 pKa = 6.56TQWARR229 pKa = 11.84SLGRR233 pKa = 11.84HH234 pKa = 5.29NYY236 pKa = 7.43FTGSLSFLDD245 pKa = 3.88YY246 pKa = 11.47DD247 pKa = 5.47DD248 pKa = 4.8FALYY252 pKa = 10.64NVIDD256 pKa = 4.96DD257 pKa = 4.16IEE259 pKa = 4.33YY260 pKa = 10.95EE261 pKa = 4.34KK262 pKa = 10.76ISTSWFKK269 pKa = 11.37SLLGAQKK276 pKa = 10.67DD277 pKa = 3.7IILKK281 pKa = 9.29GKK283 pKa = 8.39YY284 pKa = 8.83MKK286 pKa = 10.46DD287 pKa = 3.32FRR289 pKa = 11.84ISGGIPCIVLVNEE302 pKa = 4.06DD303 pKa = 3.69MDD305 pKa = 3.95WVVRR309 pKa = 11.84MASTLKK315 pKa = 10.26PWFDD319 pKa = 4.71DD320 pKa = 3.66NVLVYY325 pKa = 10.71YY326 pKa = 9.85MSASDD331 pKa = 4.0KK332 pKa = 10.91FYY334 pKa = 11.37
Molecular weight: 38.72 kDa
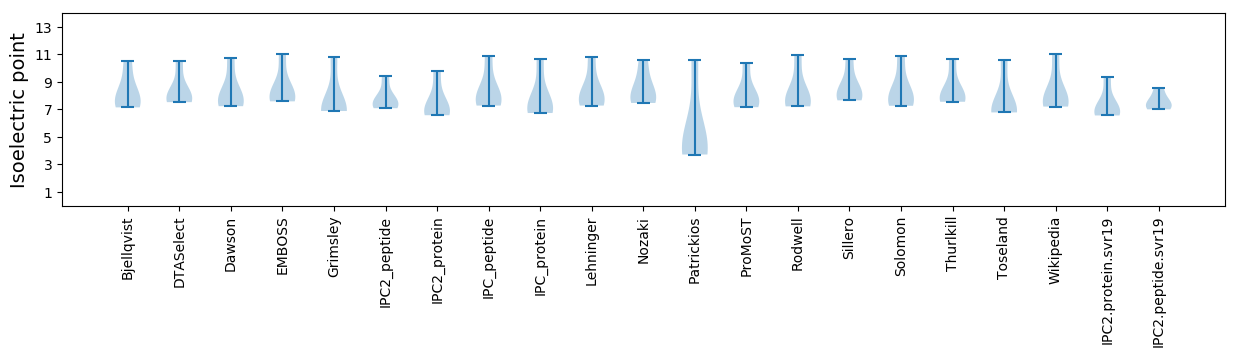
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D5BV09|A0A0D5BV09_9GEMI Capsid protein OS=Mulberry mosaic dwarf associated virus OX=1631303 PE=3 SV=1
MM1 pKa = 7.46VITRR5 pKa = 11.84SSALRR10 pKa = 11.84SRR12 pKa = 11.84RR13 pKa = 11.84GWNPGTIPRR22 pKa = 11.84RR23 pKa = 11.84PRR25 pKa = 11.84TVRR28 pKa = 11.84VTRR31 pKa = 11.84VASAMRR37 pKa = 11.84TPWRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84GPRR46 pKa = 11.84SSLARR51 pKa = 11.84RR52 pKa = 11.84KK53 pKa = 9.49RR54 pKa = 11.84RR55 pKa = 11.84PINALGPVKK64 pKa = 10.48RR65 pKa = 11.84GTFGIKK71 pKa = 9.51LASINHH77 pKa = 5.99TGGNGYY83 pKa = 9.56HH84 pKa = 6.14LTHH87 pKa = 7.21FDD89 pKa = 4.44HH90 pKa = 7.7GDD92 pKa = 3.38DD93 pKa = 3.61TNQRR97 pKa = 11.84TGRR100 pKa = 11.84KK101 pKa = 8.85IKK103 pKa = 10.6INGINIRR110 pKa = 11.84GKK112 pKa = 10.76LYY114 pKa = 10.84LNNPRR119 pKa = 11.84TNSYY123 pKa = 10.5HH124 pKa = 7.26IVRR127 pKa = 11.84LWIIRR132 pKa = 11.84DD133 pKa = 3.63TRR135 pKa = 11.84PGSEE139 pKa = 3.84PVSFSSFMDD148 pKa = 3.23MHH150 pKa = 7.47DD151 pKa = 4.42NEE153 pKa = 5.02PMTAMVKK160 pKa = 10.18KK161 pKa = 10.37DD162 pKa = 2.82WGEE165 pKa = 3.75RR166 pKa = 11.84FQVLKK171 pKa = 10.82DD172 pKa = 3.56LTFHH176 pKa = 7.1LVGANGLSFNEE187 pKa = 4.44DD188 pKa = 3.04VVEE191 pKa = 4.55EE192 pKa = 4.12YY193 pKa = 10.88FKK195 pKa = 11.28FKK197 pKa = 10.84GYY199 pKa = 10.67VLYY202 pKa = 10.79NHH204 pKa = 7.07EE205 pKa = 5.1DD206 pKa = 3.52SGSLANVLEE215 pKa = 4.23NGIFLYY221 pKa = 10.7AATSHH226 pKa = 6.48PSEE229 pKa = 4.32NVTLTANCRR238 pKa = 11.84VYY240 pKa = 10.61FYY242 pKa = 10.91DD243 pKa = 5.07AEE245 pKa = 4.19
MM1 pKa = 7.46VITRR5 pKa = 11.84SSALRR10 pKa = 11.84SRR12 pKa = 11.84RR13 pKa = 11.84GWNPGTIPRR22 pKa = 11.84RR23 pKa = 11.84PRR25 pKa = 11.84TVRR28 pKa = 11.84VTRR31 pKa = 11.84VASAMRR37 pKa = 11.84TPWRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84GPRR46 pKa = 11.84SSLARR51 pKa = 11.84RR52 pKa = 11.84KK53 pKa = 9.49RR54 pKa = 11.84RR55 pKa = 11.84PINALGPVKK64 pKa = 10.48RR65 pKa = 11.84GTFGIKK71 pKa = 9.51LASINHH77 pKa = 5.99TGGNGYY83 pKa = 9.56HH84 pKa = 6.14LTHH87 pKa = 7.21FDD89 pKa = 4.44HH90 pKa = 7.7GDD92 pKa = 3.38DD93 pKa = 3.61TNQRR97 pKa = 11.84TGRR100 pKa = 11.84KK101 pKa = 8.85IKK103 pKa = 10.6INGINIRR110 pKa = 11.84GKK112 pKa = 10.76LYY114 pKa = 10.84LNNPRR119 pKa = 11.84TNSYY123 pKa = 10.5HH124 pKa = 7.26IVRR127 pKa = 11.84LWIIRR132 pKa = 11.84DD133 pKa = 3.63TRR135 pKa = 11.84PGSEE139 pKa = 3.84PVSFSSFMDD148 pKa = 3.23MHH150 pKa = 7.47DD151 pKa = 4.42NEE153 pKa = 5.02PMTAMVKK160 pKa = 10.18KK161 pKa = 10.37DD162 pKa = 2.82WGEE165 pKa = 3.75RR166 pKa = 11.84FQVLKK171 pKa = 10.82DD172 pKa = 3.56LTFHH176 pKa = 7.1LVGANGLSFNEE187 pKa = 4.44DD188 pKa = 3.04VVEE191 pKa = 4.55EE192 pKa = 4.12YY193 pKa = 10.88FKK195 pKa = 11.28FKK197 pKa = 10.84GYY199 pKa = 10.67VLYY202 pKa = 10.79NHH204 pKa = 7.07EE205 pKa = 5.1DD206 pKa = 3.52SGSLANVLEE215 pKa = 4.23NGIFLYY221 pKa = 10.7AATSHH226 pKa = 6.48PSEE229 pKa = 4.32NVTLTANCRR238 pKa = 11.84VYY240 pKa = 10.61FYY242 pKa = 10.91DD243 pKa = 5.07AEE245 pKa = 4.19
Molecular weight: 28.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1097 |
105 |
334 |
219.4 |
25.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.108 ± 0.329 | 2.005 ± 0.599 |
5.287 ± 0.464 | 5.834 ± 0.541 |
4.74 ± 0.516 | 5.287 ± 0.724 |
3.373 ± 0.345 | 5.378 ± 0.494 |
5.287 ± 0.541 | 7.475 ± 0.225 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.097 ± 0.311 | 4.74 ± 0.652 |
6.016 ± 0.655 | 3.464 ± 0.819 |
7.84 ± 0.869 | 8.204 ± 0.358 |
5.014 ± 0.533 | 5.925 ± 0.206 |
2.005 ± 0.314 | 3.92 ± 0.621 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
