
Big Cypress virus
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.1
Get precalculated fractions of proteins
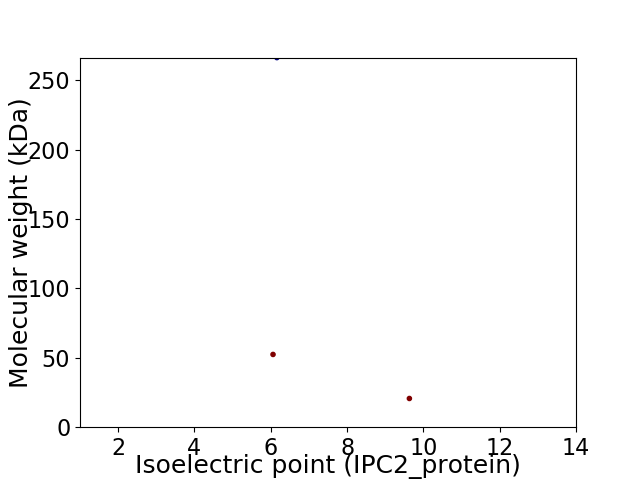
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
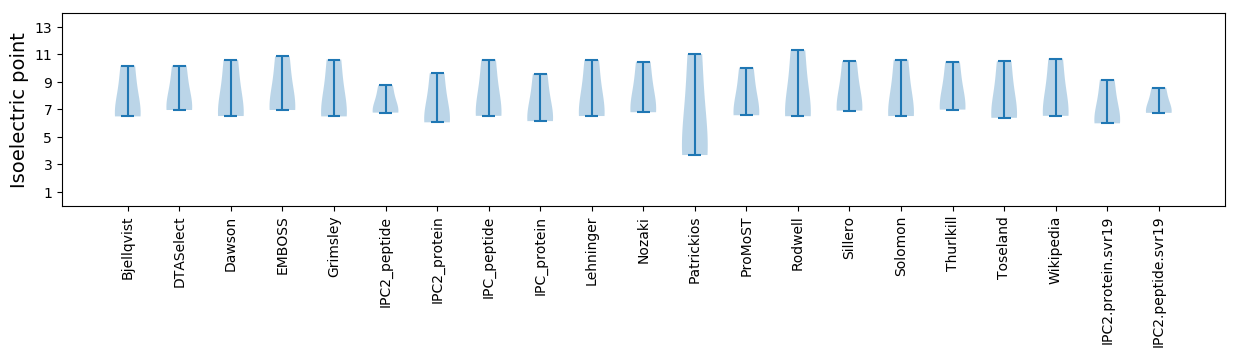
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1U7EIT7|A0A1U7EIT7_9VIRU Uncharacterized protein OS=Big Cypress virus OX=1955196 PE=4 SV=1
MM1 pKa = 7.8EE2 pKa = 4.46FTKK5 pKa = 10.45TSQDD9 pKa = 3.28FLLQSLLCVSAILSIPFFTLNTLTEE34 pKa = 4.1LTTRR38 pKa = 11.84SLSTIISINLMINHH52 pKa = 7.19ISLIMNFFLLFCLSAATASVLRR74 pKa = 11.84NEE76 pKa = 5.56LINIIKK82 pKa = 10.5ARR84 pKa = 11.84DD85 pKa = 3.68VPDD88 pKa = 4.64GLPKK92 pKa = 10.54SQIRR96 pKa = 11.84TNLRR100 pKa = 11.84IMRR103 pKa = 11.84AFPNFILSNEE113 pKa = 4.21QTVHH117 pKa = 6.95DD118 pKa = 4.7SPVTFDD124 pKa = 3.92KK125 pKa = 11.12EE126 pKa = 3.98CLEE129 pKa = 4.06AHH131 pKa = 5.81STQSFLFVGCNLPKK145 pKa = 9.78HH146 pKa = 6.22CKK148 pKa = 9.93NVVTHH153 pKa = 6.79TSDD156 pKa = 2.97HH157 pKa = 6.31SLLGFEE163 pKa = 5.27SVYY166 pKa = 10.76CLSFHH171 pKa = 6.79QSNDD175 pKa = 2.92SKK177 pKa = 11.12QISLFDD183 pKa = 3.82LSFEE187 pKa = 3.99KK188 pKa = 10.57RR189 pKa = 11.84DD190 pKa = 3.4IKK192 pKa = 10.24FTAEE196 pKa = 3.65FDD198 pKa = 3.79FQPRR202 pKa = 11.84TTNILKK208 pKa = 10.22NAIPVSFFEE217 pKa = 4.45NFFIAKK223 pKa = 8.43TEE225 pKa = 3.92GRR227 pKa = 11.84FYY229 pKa = 11.25LLPRR233 pKa = 11.84VCRR236 pKa = 11.84EE237 pKa = 3.54RR238 pKa = 11.84FTSDD242 pKa = 2.24SHH244 pKa = 6.14YY245 pKa = 11.03VPSTFTLKK253 pKa = 9.96TDD255 pKa = 3.74DD256 pKa = 3.67VSFCIAHH263 pKa = 6.64KK264 pKa = 10.39FFSNVDD270 pKa = 3.42CTLDD274 pKa = 3.36VGFVSMYY281 pKa = 11.32DD282 pKa = 3.16LFDD285 pKa = 4.24YY286 pKa = 11.46YY287 pKa = 10.94DD288 pKa = 4.0FPVSYY293 pKa = 10.71DD294 pKa = 3.56GAPGAAYY301 pKa = 10.27SGNNIDD307 pKa = 3.4QTEE310 pKa = 4.25FRR312 pKa = 11.84AEE314 pKa = 3.79QYY316 pKa = 10.16YY317 pKa = 10.34KK318 pKa = 10.66SHH320 pKa = 6.8AADD323 pKa = 4.79FIYY326 pKa = 10.05PVSLRR331 pKa = 11.84KK332 pKa = 10.26GGTYY336 pKa = 10.73LSNTKK341 pKa = 10.13SPYY344 pKa = 7.06PPKK347 pKa = 10.79YY348 pKa = 10.09IVFSKK353 pKa = 11.09NDD355 pKa = 3.16ISSTTVHH362 pKa = 6.46CFPRR366 pKa = 11.84LSHH369 pKa = 6.0YY370 pKa = 10.87KK371 pKa = 9.97PFLSTVLSSFLAEE384 pKa = 4.25IGGLFQKK391 pKa = 10.56IFDD394 pKa = 4.11TLIEE398 pKa = 4.28CSEE401 pKa = 4.49TIAEE405 pKa = 5.22FILKK409 pKa = 10.28LLGHH413 pKa = 5.86VVSKK417 pKa = 9.72FLNALLPYY425 pKa = 9.72LSPRR429 pKa = 11.84IVPPLLAFPVAYY441 pKa = 10.07FCFSDD446 pKa = 4.04VIKK449 pKa = 10.93SVIFSFFVFICVV461 pKa = 3.14
MM1 pKa = 7.8EE2 pKa = 4.46FTKK5 pKa = 10.45TSQDD9 pKa = 3.28FLLQSLLCVSAILSIPFFTLNTLTEE34 pKa = 4.1LTTRR38 pKa = 11.84SLSTIISINLMINHH52 pKa = 7.19ISLIMNFFLLFCLSAATASVLRR74 pKa = 11.84NEE76 pKa = 5.56LINIIKK82 pKa = 10.5ARR84 pKa = 11.84DD85 pKa = 3.68VPDD88 pKa = 4.64GLPKK92 pKa = 10.54SQIRR96 pKa = 11.84TNLRR100 pKa = 11.84IMRR103 pKa = 11.84AFPNFILSNEE113 pKa = 4.21QTVHH117 pKa = 6.95DD118 pKa = 4.7SPVTFDD124 pKa = 3.92KK125 pKa = 11.12EE126 pKa = 3.98CLEE129 pKa = 4.06AHH131 pKa = 5.81STQSFLFVGCNLPKK145 pKa = 9.78HH146 pKa = 6.22CKK148 pKa = 9.93NVVTHH153 pKa = 6.79TSDD156 pKa = 2.97HH157 pKa = 6.31SLLGFEE163 pKa = 5.27SVYY166 pKa = 10.76CLSFHH171 pKa = 6.79QSNDD175 pKa = 2.92SKK177 pKa = 11.12QISLFDD183 pKa = 3.82LSFEE187 pKa = 3.99KK188 pKa = 10.57RR189 pKa = 11.84DD190 pKa = 3.4IKK192 pKa = 10.24FTAEE196 pKa = 3.65FDD198 pKa = 3.79FQPRR202 pKa = 11.84TTNILKK208 pKa = 10.22NAIPVSFFEE217 pKa = 4.45NFFIAKK223 pKa = 8.43TEE225 pKa = 3.92GRR227 pKa = 11.84FYY229 pKa = 11.25LLPRR233 pKa = 11.84VCRR236 pKa = 11.84EE237 pKa = 3.54RR238 pKa = 11.84FTSDD242 pKa = 2.24SHH244 pKa = 6.14YY245 pKa = 11.03VPSTFTLKK253 pKa = 9.96TDD255 pKa = 3.74DD256 pKa = 3.67VSFCIAHH263 pKa = 6.64KK264 pKa = 10.39FFSNVDD270 pKa = 3.42CTLDD274 pKa = 3.36VGFVSMYY281 pKa = 11.32DD282 pKa = 3.16LFDD285 pKa = 4.24YY286 pKa = 11.46YY287 pKa = 10.94DD288 pKa = 4.0FPVSYY293 pKa = 10.71DD294 pKa = 3.56GAPGAAYY301 pKa = 10.27SGNNIDD307 pKa = 3.4QTEE310 pKa = 4.25FRR312 pKa = 11.84AEE314 pKa = 3.79QYY316 pKa = 10.16YY317 pKa = 10.34KK318 pKa = 10.66SHH320 pKa = 6.8AADD323 pKa = 4.79FIYY326 pKa = 10.05PVSLRR331 pKa = 11.84KK332 pKa = 10.26GGTYY336 pKa = 10.73LSNTKK341 pKa = 10.13SPYY344 pKa = 7.06PPKK347 pKa = 10.79YY348 pKa = 10.09IVFSKK353 pKa = 11.09NDD355 pKa = 3.16ISSTTVHH362 pKa = 6.46CFPRR366 pKa = 11.84LSHH369 pKa = 6.0YY370 pKa = 10.87KK371 pKa = 9.97PFLSTVLSSFLAEE384 pKa = 4.25IGGLFQKK391 pKa = 10.56IFDD394 pKa = 4.11TLIEE398 pKa = 4.28CSEE401 pKa = 4.49TIAEE405 pKa = 5.22FILKK409 pKa = 10.28LLGHH413 pKa = 5.86VVSKK417 pKa = 9.72FLNALLPYY425 pKa = 9.72LSPRR429 pKa = 11.84IVPPLLAFPVAYY441 pKa = 10.07FCFSDD446 pKa = 4.04VIKK449 pKa = 10.93SVIFSFFVFICVV461 pKa = 3.14
Molecular weight: 52.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1U7EIT7|A0A1U7EIT7_9VIRU Uncharacterized protein OS=Big Cypress virus OX=1955196 PE=4 SV=1
MM1 pKa = 7.81SSNQKK6 pKa = 9.83NSKK9 pKa = 8.99PRR11 pKa = 11.84QRR13 pKa = 11.84KK14 pKa = 6.59QVVYY18 pKa = 8.91KK19 pKa = 9.66TVQTRR24 pKa = 11.84TPAKK28 pKa = 10.02LKK30 pKa = 10.45KK31 pKa = 10.13SPSKK35 pKa = 10.21KK36 pKa = 5.25TTKK39 pKa = 8.35WTIDD43 pKa = 3.22SGFTDD48 pKa = 3.62ILNSFTSSVSKK59 pKa = 10.12PAVLLSIIAISAFVVTHH76 pKa = 6.41SSSFSTSIVGKK87 pKa = 8.83WANDD91 pKa = 3.64HH92 pKa = 6.66KK93 pKa = 10.88NTTIGKK99 pKa = 7.77WVSEE103 pKa = 4.05NDD105 pKa = 3.35EE106 pKa = 4.46KK107 pKa = 11.62LLGLAVFAPAVLNSPNSIRR126 pKa = 11.84VMLILCSFLWIVLIPEE142 pKa = 3.95NSVYY146 pKa = 10.27QYY148 pKa = 11.05CLQSLALHH156 pKa = 6.72TYY158 pKa = 9.88FKK160 pKa = 10.98VRR162 pKa = 11.84NPKK165 pKa = 10.17SRR167 pKa = 11.84FLVIAVVVIAYY178 pKa = 9.21IAGFLVVKK186 pKa = 10.62
MM1 pKa = 7.81SSNQKK6 pKa = 9.83NSKK9 pKa = 8.99PRR11 pKa = 11.84QRR13 pKa = 11.84KK14 pKa = 6.59QVVYY18 pKa = 8.91KK19 pKa = 9.66TVQTRR24 pKa = 11.84TPAKK28 pKa = 10.02LKK30 pKa = 10.45KK31 pKa = 10.13SPSKK35 pKa = 10.21KK36 pKa = 5.25TTKK39 pKa = 8.35WTIDD43 pKa = 3.22SGFTDD48 pKa = 3.62ILNSFTSSVSKK59 pKa = 10.12PAVLLSIIAISAFVVTHH76 pKa = 6.41SSSFSTSIVGKK87 pKa = 8.83WANDD91 pKa = 3.64HH92 pKa = 6.66KK93 pKa = 10.88NTTIGKK99 pKa = 7.77WVSEE103 pKa = 4.05NDD105 pKa = 3.35EE106 pKa = 4.46KK107 pKa = 11.62LLGLAVFAPAVLNSPNSIRR126 pKa = 11.84VMLILCSFLWIVLIPEE142 pKa = 3.95NSVYY146 pKa = 10.27QYY148 pKa = 11.05CLQSLALHH156 pKa = 6.72TYY158 pKa = 9.88FKK160 pKa = 10.98VRR162 pKa = 11.84NPKK165 pKa = 10.17SRR167 pKa = 11.84FLVIAVVVIAYY178 pKa = 9.21IAGFLVVKK186 pKa = 10.62
Molecular weight: 20.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2980 |
186 |
2333 |
993.3 |
113.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.671 ± 0.307 | 1.946 ± 0.298 |
5.671 ± 0.727 | 5.57 ± 1.137 |
7.349 ± 1.123 | 3.591 ± 0.39 |
2.517 ± 0.153 | 6.577 ± 0.332 |
6.779 ± 0.652 | 8.993 ± 0.713 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.611 ± 0.256 | 6.208 ± 0.704 |
4.128 ± 0.341 | 2.617 ± 0.106 |
4.497 ± 0.529 | 8.893 ± 1.168 |
6.678 ± 0.134 | 6.477 ± 0.837 |
0.537 ± 0.273 | 3.691 ± 0.181 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
