
Puniceispirillum phage HMO-2011
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; unclassified Podoviridae
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
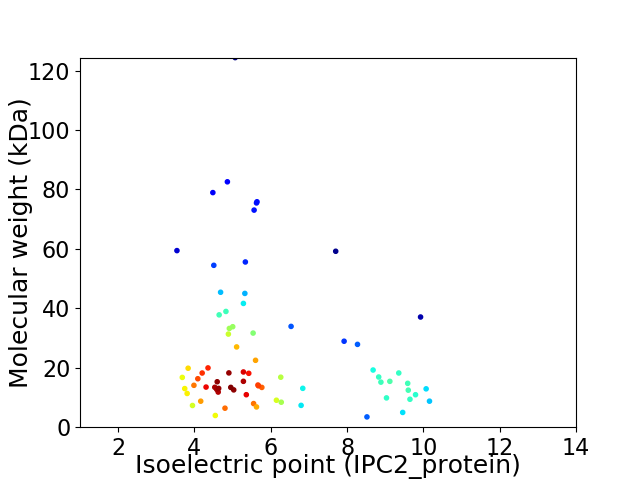
Virtual 2D-PAGE plot for 74 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
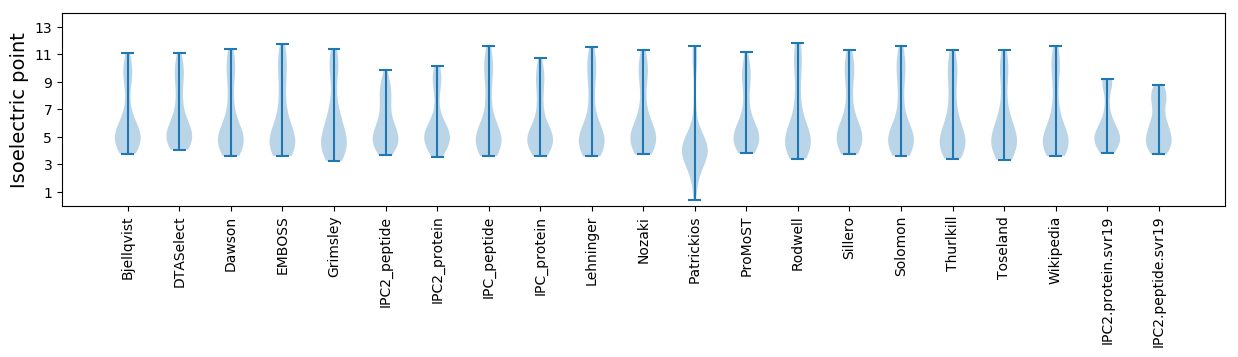
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S4S2M4|S4S2M4_9CAUD Putative tail fiber protein OS=Puniceispirillum phage HMO-2011 OX=948071 GN=phage1322_40 PE=4 SV=1
MM1 pKa = 7.87PYY3 pKa = 9.68IGKK6 pKa = 9.45SPQNGVRR13 pKa = 11.84NRR15 pKa = 11.84FIYY18 pKa = 10.12QATAGQTSFSGSDD31 pKa = 3.23SDD33 pKa = 5.25SKK35 pKa = 11.54VLTYY39 pKa = 10.52QDD41 pKa = 3.89GLYY44 pKa = 9.76MDD46 pKa = 4.73VFQNGVLLKK55 pKa = 10.64PGTDD59 pKa = 3.62YY60 pKa = 10.63TATTGTTVVLVTGASSGDD78 pKa = 3.64VVEE81 pKa = 4.66MVAYY85 pKa = 10.28DD86 pKa = 3.78VFSVANSYY94 pKa = 8.3TVTEE98 pKa = 4.02SDD100 pKa = 2.54TRR102 pKa = 11.84YY103 pKa = 9.72PFKK106 pKa = 10.99GNNSIIRR113 pKa = 11.84LNGQSITSDD122 pKa = 3.24VTIDD126 pKa = 3.35ADD128 pKa = 4.06EE129 pKa = 5.04NGVSAGPITQDD140 pKa = 2.94NATVTVNGYY149 pKa = 8.79WSIVV153 pKa = 3.22
MM1 pKa = 7.87PYY3 pKa = 9.68IGKK6 pKa = 9.45SPQNGVRR13 pKa = 11.84NRR15 pKa = 11.84FIYY18 pKa = 10.12QATAGQTSFSGSDD31 pKa = 3.23SDD33 pKa = 5.25SKK35 pKa = 11.54VLTYY39 pKa = 10.52QDD41 pKa = 3.89GLYY44 pKa = 9.76MDD46 pKa = 4.73VFQNGVLLKK55 pKa = 10.64PGTDD59 pKa = 3.62YY60 pKa = 10.63TATTGTTVVLVTGASSGDD78 pKa = 3.64VVEE81 pKa = 4.66MVAYY85 pKa = 10.28DD86 pKa = 3.78VFSVANSYY94 pKa = 8.3TVTEE98 pKa = 4.02SDD100 pKa = 2.54TRR102 pKa = 11.84YY103 pKa = 9.72PFKK106 pKa = 10.99GNNSIIRR113 pKa = 11.84LNGQSITSDD122 pKa = 3.24VTIDD126 pKa = 3.35ADD128 pKa = 4.06EE129 pKa = 5.04NGVSAGPITQDD140 pKa = 2.94NATVTVNGYY149 pKa = 8.79WSIVV153 pKa = 3.22
Molecular weight: 16.31 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S4S2G2|S4S2G2_9CAUD Uncharacterized protein OS=Puniceispirillum phage HMO-2011 OX=948071 GN=phage1322_73 PE=4 SV=1
MM1 pKa = 7.2TRR3 pKa = 11.84INLFALDD10 pKa = 3.66SKK12 pKa = 11.56AYY14 pKa = 9.93LVLGRR19 pKa = 11.84TLSLLKK25 pKa = 9.98IKK27 pKa = 10.25HH28 pKa = 5.21VRR30 pKa = 11.84SPTMHH35 pKa = 7.0VMMHH39 pKa = 6.53RR40 pKa = 11.84GSRR43 pKa = 11.84PASCQRR49 pKa = 11.84STGHH53 pKa = 6.46IKK55 pKa = 11.01LNGNIGDD62 pKa = 3.46ISMGIIKK69 pKa = 8.84TLAKK73 pKa = 10.25GAAKK77 pKa = 10.07KK78 pKa = 10.37ARR80 pKa = 11.84EE81 pKa = 4.05KK82 pKa = 10.78QGEE85 pKa = 3.94AGKK88 pKa = 10.52ARR90 pKa = 11.84RR91 pKa = 11.84AAKK94 pKa = 10.29AKK96 pKa = 10.83AEE98 pKa = 4.07AAKK101 pKa = 10.35KK102 pKa = 9.93AASKK106 pKa = 10.4KK107 pKa = 10.13AASKK111 pKa = 10.55PKK113 pKa = 9.23ATQKK117 pKa = 10.93GLRR120 pKa = 11.84LSRR123 pKa = 11.84KK124 pKa = 8.71PVTQEE129 pKa = 3.97GMGLDD134 pKa = 4.6AITGRR139 pKa = 11.84SNMAAASVDD148 pKa = 3.06KK149 pKa = 11.2GKK151 pKa = 10.57AGKK154 pKa = 7.59VTRR157 pKa = 11.84SAEE160 pKa = 3.95PGFLQSQRR168 pKa = 11.84TAGSRR173 pKa = 11.84AKK175 pKa = 10.47AQEE178 pKa = 4.06KK179 pKa = 10.42VDD181 pKa = 3.94LAKK184 pKa = 10.48KK185 pKa = 10.01VRR187 pKa = 11.84DD188 pKa = 3.81GKK190 pKa = 9.46ATKK193 pKa = 10.37AEE195 pKa = 4.06KK196 pKa = 10.6AKK198 pKa = 10.76LKK200 pKa = 10.41SLRR203 pKa = 11.84EE204 pKa = 3.81KK205 pKa = 10.45DD206 pKa = 3.86AKK208 pKa = 10.07DD209 pKa = 3.23TASARR214 pKa = 11.84AKK216 pKa = 10.2GAASRR221 pKa = 11.84KK222 pKa = 8.77KK223 pKa = 10.09KK224 pKa = 10.76SKK226 pKa = 9.59ATLPEE231 pKa = 4.43LKK233 pKa = 10.05SAPSKK238 pKa = 10.79KK239 pKa = 8.28VTKK242 pKa = 9.79EE243 pKa = 3.65KK244 pKa = 10.8PPINLKK250 pKa = 9.43TGEE253 pKa = 4.1INKK256 pKa = 9.88SVFNKK261 pKa = 10.33LSPGKK266 pKa = 9.96QEE268 pKa = 3.81QAVRR272 pKa = 11.84NAMARR277 pKa = 11.84VEE279 pKa = 4.35GPRR282 pKa = 11.84KK283 pKa = 9.74RR284 pKa = 11.84EE285 pKa = 3.6LKK287 pKa = 10.82AFLEE291 pKa = 4.48EE292 pKa = 4.1MRR294 pKa = 11.84KK295 pKa = 9.64SKK297 pKa = 10.49PGEE300 pKa = 3.79SGLRR304 pKa = 11.84RR305 pKa = 11.84RR306 pKa = 11.84KK307 pKa = 8.31GTRR310 pKa = 11.84GMSGAEE316 pKa = 3.93SSIKK320 pKa = 10.75DD321 pKa = 3.4LDD323 pKa = 3.75KK324 pKa = 11.48GVGGRR329 pKa = 11.84GGLDD333 pKa = 3.43FNRR336 pKa = 11.84GGMVKK341 pKa = 9.91KK342 pKa = 10.38RR343 pKa = 11.84KK344 pKa = 9.29KK345 pKa = 10.24
MM1 pKa = 7.2TRR3 pKa = 11.84INLFALDD10 pKa = 3.66SKK12 pKa = 11.56AYY14 pKa = 9.93LVLGRR19 pKa = 11.84TLSLLKK25 pKa = 9.98IKK27 pKa = 10.25HH28 pKa = 5.21VRR30 pKa = 11.84SPTMHH35 pKa = 7.0VMMHH39 pKa = 6.53RR40 pKa = 11.84GSRR43 pKa = 11.84PASCQRR49 pKa = 11.84STGHH53 pKa = 6.46IKK55 pKa = 11.01LNGNIGDD62 pKa = 3.46ISMGIIKK69 pKa = 8.84TLAKK73 pKa = 10.25GAAKK77 pKa = 10.07KK78 pKa = 10.37ARR80 pKa = 11.84EE81 pKa = 4.05KK82 pKa = 10.78QGEE85 pKa = 3.94AGKK88 pKa = 10.52ARR90 pKa = 11.84RR91 pKa = 11.84AAKK94 pKa = 10.29AKK96 pKa = 10.83AEE98 pKa = 4.07AAKK101 pKa = 10.35KK102 pKa = 9.93AASKK106 pKa = 10.4KK107 pKa = 10.13AASKK111 pKa = 10.55PKK113 pKa = 9.23ATQKK117 pKa = 10.93GLRR120 pKa = 11.84LSRR123 pKa = 11.84KK124 pKa = 8.71PVTQEE129 pKa = 3.97GMGLDD134 pKa = 4.6AITGRR139 pKa = 11.84SNMAAASVDD148 pKa = 3.06KK149 pKa = 11.2GKK151 pKa = 10.57AGKK154 pKa = 7.59VTRR157 pKa = 11.84SAEE160 pKa = 3.95PGFLQSQRR168 pKa = 11.84TAGSRR173 pKa = 11.84AKK175 pKa = 10.47AQEE178 pKa = 4.06KK179 pKa = 10.42VDD181 pKa = 3.94LAKK184 pKa = 10.48KK185 pKa = 10.01VRR187 pKa = 11.84DD188 pKa = 3.81GKK190 pKa = 9.46ATKK193 pKa = 10.37AEE195 pKa = 4.06KK196 pKa = 10.6AKK198 pKa = 10.76LKK200 pKa = 10.41SLRR203 pKa = 11.84EE204 pKa = 3.81KK205 pKa = 10.45DD206 pKa = 3.86AKK208 pKa = 10.07DD209 pKa = 3.23TASARR214 pKa = 11.84AKK216 pKa = 10.2GAASRR221 pKa = 11.84KK222 pKa = 8.77KK223 pKa = 10.09KK224 pKa = 10.76SKK226 pKa = 9.59ATLPEE231 pKa = 4.43LKK233 pKa = 10.05SAPSKK238 pKa = 10.79KK239 pKa = 8.28VTKK242 pKa = 9.79EE243 pKa = 3.65KK244 pKa = 10.8PPINLKK250 pKa = 9.43TGEE253 pKa = 4.1INKK256 pKa = 9.88SVFNKK261 pKa = 10.33LSPGKK266 pKa = 9.96QEE268 pKa = 3.81QAVRR272 pKa = 11.84NAMARR277 pKa = 11.84VEE279 pKa = 4.35GPRR282 pKa = 11.84KK283 pKa = 9.74RR284 pKa = 11.84EE285 pKa = 3.6LKK287 pKa = 10.82AFLEE291 pKa = 4.48EE292 pKa = 4.1MRR294 pKa = 11.84KK295 pKa = 9.64SKK297 pKa = 10.49PGEE300 pKa = 3.79SGLRR304 pKa = 11.84RR305 pKa = 11.84RR306 pKa = 11.84KK307 pKa = 8.31GTRR310 pKa = 11.84GMSGAEE316 pKa = 3.93SSIKK320 pKa = 10.75DD321 pKa = 3.4LDD323 pKa = 3.75KK324 pKa = 11.48GVGGRR329 pKa = 11.84GGLDD333 pKa = 3.43FNRR336 pKa = 11.84GGMVKK341 pKa = 9.91KK342 pKa = 10.38RR343 pKa = 11.84KK344 pKa = 9.29KK345 pKa = 10.24
Molecular weight: 37.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
17041 |
30 |
1128 |
230.3 |
25.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.536 ± 0.495 | 0.78 ± 0.113 |
6.76 ± 0.279 | 6.109 ± 0.44 |
3.433 ± 0.208 | 7.306 ± 0.343 |
1.491 ± 0.194 | 5.047 ± 0.179 |
6.238 ± 0.504 | 7.153 ± 0.249 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.163 ± 0.232 | 4.841 ± 0.21 |
3.767 ± 0.267 | 4.178 ± 0.35 |
4.853 ± 0.305 | 6.754 ± 0.356 |
7.288 ± 0.497 | 6.602 ± 0.252 |
1.262 ± 0.138 | 3.439 ± 0.174 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
