
Human rotavirus C
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Rotavirus; Rotavirus C
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
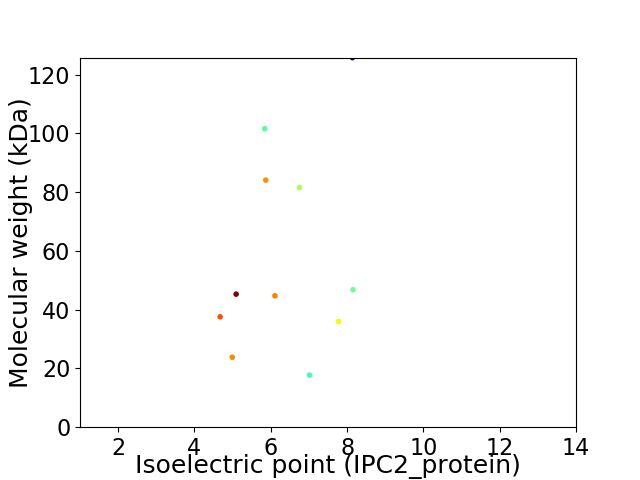
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D0VZZ4|D0VZZ4_9REOV Outer capsid protein VP4 OS=Human rotavirus C OX=10943 GN=VP4 PE=3 SV=2
MM1 pKa = 7.33VCTTLYY7 pKa = 8.12TVCVILFILFIYY19 pKa = 10.33ILLFRR24 pKa = 11.84KK25 pKa = 8.46MFHH28 pKa = 7.25LITDD32 pKa = 4.06TLIVMLILSNCVEE45 pKa = 4.35WSQGQMFIDD54 pKa = 4.65DD55 pKa = 4.11IYY57 pKa = 11.2YY58 pKa = 10.05NGNVEE63 pKa = 4.41TIINSTDD70 pKa = 3.42PFDD73 pKa = 4.46VEE75 pKa = 4.36SLCIYY80 pKa = 9.75FPNAVVGSQGPGKK93 pKa = 10.4SDD95 pKa = 2.77GHH97 pKa = 8.08LNDD100 pKa = 3.7GNYY103 pKa = 10.46AQTIATLFEE112 pKa = 4.43TKK114 pKa = 10.21GFPKK118 pKa = 10.58GSIILKK124 pKa = 9.46TYY126 pKa = 7.58TQTSDD131 pKa = 4.08FINSVEE137 pKa = 4.26MTCSYY142 pKa = 10.97NIVIIPDD149 pKa = 3.7SPNDD153 pKa = 3.66SEE155 pKa = 5.39SIEE158 pKa = 4.82QIAEE162 pKa = 3.94WILNVWRR169 pKa = 11.84CDD171 pKa = 3.57DD172 pKa = 4.06MNLEE176 pKa = 4.3IYY178 pKa = 9.64TYY180 pKa = 9.6EE181 pKa = 3.99QIGINNLWAAFGSDD195 pKa = 4.29CDD197 pKa = 4.41ISVCPLDD204 pKa = 3.68TTSNGIGCSPASTEE218 pKa = 4.12TYY220 pKa = 10.02EE221 pKa = 4.47VVSNDD226 pKa = 3.33TQLTLINVVDD236 pKa = 4.05NVRR239 pKa = 11.84HH240 pKa = 6.38RR241 pKa = 11.84IQMNTAQCKK250 pKa = 9.81LKK252 pKa = 10.97NCIKK256 pKa = 10.93GEE258 pKa = 3.91ARR260 pKa = 11.84LNTALIRR267 pKa = 11.84ISTSSSFDD275 pKa = 3.34NSLSPLNNGQTTRR288 pKa = 11.84SFKK291 pKa = 10.82INAKK295 pKa = 9.15KK296 pKa = 9.06WWTIFYY302 pKa = 9.29TIIDD306 pKa = 4.45YY307 pKa = 11.08INTIVQAMTPRR318 pKa = 11.84HH319 pKa = 5.18RR320 pKa = 11.84AIYY323 pKa = 9.33PEE325 pKa = 3.79GWMLRR330 pKa = 11.84YY331 pKa = 9.45AA332 pKa = 4.23
MM1 pKa = 7.33VCTTLYY7 pKa = 8.12TVCVILFILFIYY19 pKa = 10.33ILLFRR24 pKa = 11.84KK25 pKa = 8.46MFHH28 pKa = 7.25LITDD32 pKa = 4.06TLIVMLILSNCVEE45 pKa = 4.35WSQGQMFIDD54 pKa = 4.65DD55 pKa = 4.11IYY57 pKa = 11.2YY58 pKa = 10.05NGNVEE63 pKa = 4.41TIINSTDD70 pKa = 3.42PFDD73 pKa = 4.46VEE75 pKa = 4.36SLCIYY80 pKa = 9.75FPNAVVGSQGPGKK93 pKa = 10.4SDD95 pKa = 2.77GHH97 pKa = 8.08LNDD100 pKa = 3.7GNYY103 pKa = 10.46AQTIATLFEE112 pKa = 4.43TKK114 pKa = 10.21GFPKK118 pKa = 10.58GSIILKK124 pKa = 9.46TYY126 pKa = 7.58TQTSDD131 pKa = 4.08FINSVEE137 pKa = 4.26MTCSYY142 pKa = 10.97NIVIIPDD149 pKa = 3.7SPNDD153 pKa = 3.66SEE155 pKa = 5.39SIEE158 pKa = 4.82QIAEE162 pKa = 3.94WILNVWRR169 pKa = 11.84CDD171 pKa = 3.57DD172 pKa = 4.06MNLEE176 pKa = 4.3IYY178 pKa = 9.64TYY180 pKa = 9.6EE181 pKa = 3.99QIGINNLWAAFGSDD195 pKa = 4.29CDD197 pKa = 4.41ISVCPLDD204 pKa = 3.68TTSNGIGCSPASTEE218 pKa = 4.12TYY220 pKa = 10.02EE221 pKa = 4.47VVSNDD226 pKa = 3.33TQLTLINVVDD236 pKa = 4.05NVRR239 pKa = 11.84HH240 pKa = 6.38RR241 pKa = 11.84IQMNTAQCKK250 pKa = 9.81LKK252 pKa = 10.97NCIKK256 pKa = 10.93GEE258 pKa = 3.91ARR260 pKa = 11.84LNTALIRR267 pKa = 11.84ISTSSSFDD275 pKa = 3.34NSLSPLNNGQTTRR288 pKa = 11.84SFKK291 pKa = 10.82INAKK295 pKa = 9.15KK296 pKa = 9.06WWTIFYY302 pKa = 9.29TIIDD306 pKa = 4.45YY307 pKa = 11.08INTIVQAMTPRR318 pKa = 11.84HH319 pKa = 5.18RR320 pKa = 11.84AIYY323 pKa = 9.33PEE325 pKa = 3.79GWMLRR330 pKa = 11.84YY331 pKa = 9.45AA332 pKa = 4.23
Molecular weight: 37.56 kDa
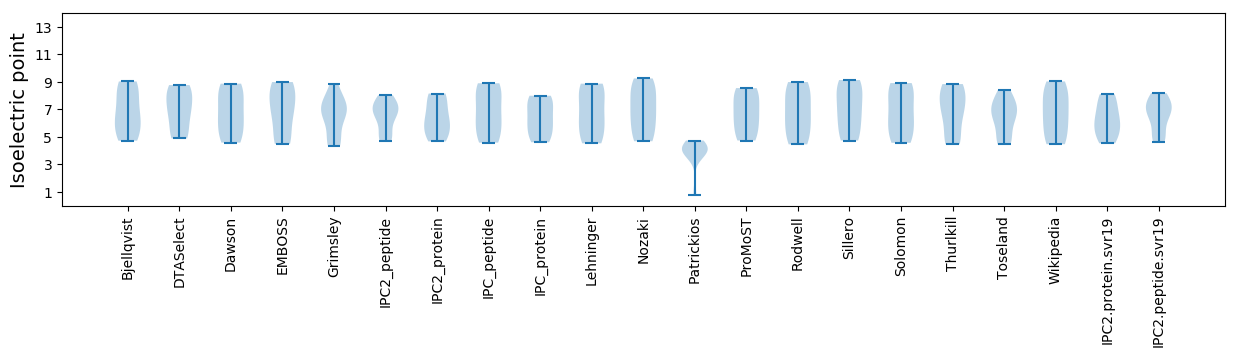
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A160NTT5|A0A160NTT5_9REOV Non-structural protein 5 OS=Human rotavirus C OX=10943 GN=NSP5 PE=3 SV=1
MM1 pKa = 7.45ANSFRR6 pKa = 11.84EE7 pKa = 3.69MLYY10 pKa = 9.54WYY12 pKa = 10.31RR13 pKa = 11.84KK14 pKa = 9.77IIDD17 pKa = 3.64RR18 pKa = 11.84KK19 pKa = 9.82LPCVNVNIWRR29 pKa = 11.84RR30 pKa = 11.84EE31 pKa = 3.69IEE33 pKa = 4.08YY34 pKa = 10.3KK35 pKa = 11.14ANGVCLNCLNEE46 pKa = 4.79CKK48 pKa = 10.41VCPCDD53 pKa = 3.46YY54 pKa = 10.75CGIRR58 pKa = 11.84HH59 pKa = 5.94KK60 pKa = 11.09CEE62 pKa = 3.44NCLNSDD68 pKa = 4.11CFMNTDD74 pKa = 3.49NEE76 pKa = 4.62FNSHH80 pKa = 5.22RR81 pKa = 11.84WITFDD86 pKa = 4.02EE87 pKa = 5.0EE88 pKa = 4.11PSQMVLFEE96 pKa = 3.83YY97 pKa = 9.86WIMYY101 pKa = 9.91KK102 pKa = 10.47DD103 pKa = 3.64YY104 pKa = 10.65FLSKK108 pKa = 10.48FNYY111 pKa = 9.45NYY113 pKa = 10.52KK114 pKa = 9.94IQLKK118 pKa = 9.07ILNMNKK124 pKa = 9.16NRR126 pKa = 11.84RR127 pKa = 11.84FHH129 pKa = 7.83INEE132 pKa = 4.12SKK134 pKa = 10.67KK135 pKa = 10.41KK136 pKa = 9.48ALSVPITSQYY146 pKa = 11.63LKK148 pKa = 10.79FKK150 pKa = 10.23FNNKK154 pKa = 8.34IYY156 pKa = 10.62IMFGTFLTSKK166 pKa = 8.75IQPWIQLKK174 pKa = 8.78SLKK177 pKa = 10.28VGYY180 pKa = 9.33IQSLNVDD187 pKa = 3.8RR188 pKa = 11.84CAKK191 pKa = 10.54LIATKK196 pKa = 10.81GMFATNSFKK205 pKa = 10.85SSCITEE211 pKa = 3.76INARR215 pKa = 11.84RR216 pKa = 11.84PISEE220 pKa = 4.25CDD222 pKa = 3.59YY223 pKa = 11.23LIEE226 pKa = 5.28ACLCNEE232 pKa = 3.59NNEE235 pKa = 4.35WKK237 pKa = 10.42FSAVMGRR244 pKa = 11.84DD245 pKa = 3.68KK246 pKa = 11.23IPVTKK251 pKa = 10.18SLAMKK256 pKa = 7.77YY257 pKa = 9.59FCKK260 pKa = 10.66NINTEE265 pKa = 3.87LFYY268 pKa = 11.19YY269 pKa = 10.18GHH271 pKa = 7.19AKK273 pKa = 9.52CHH275 pKa = 5.44VVSEE279 pKa = 4.47CPRR282 pKa = 11.84WNQQLRR288 pKa = 11.84VLHH291 pKa = 6.91ASTLNIIFRR300 pKa = 11.84RR301 pKa = 11.84QFMNEE306 pKa = 3.46VVEE309 pKa = 4.18WFEE312 pKa = 5.58NFTQLTGMHH321 pKa = 6.96HH322 pKa = 7.37DD323 pKa = 4.76FIKK326 pKa = 9.28TCVYY330 pKa = 9.97NKK332 pKa = 10.13VIVSHH337 pKa = 5.99FRR339 pKa = 11.84KK340 pKa = 10.18EE341 pKa = 3.63IEE343 pKa = 4.27DD344 pKa = 3.84YY345 pKa = 11.32INFGKK350 pKa = 10.36KK351 pKa = 9.23ISLSSVIPDD360 pKa = 2.95GHH362 pKa = 7.46ALYY365 pKa = 10.95NSIDD369 pKa = 3.66RR370 pKa = 11.84LRR372 pKa = 11.84ISLMLAIDD380 pKa = 3.56VALNRR385 pKa = 11.84IEE387 pKa = 4.11SQQMDD392 pKa = 3.91VLL394 pKa = 4.2
MM1 pKa = 7.45ANSFRR6 pKa = 11.84EE7 pKa = 3.69MLYY10 pKa = 9.54WYY12 pKa = 10.31RR13 pKa = 11.84KK14 pKa = 9.77IIDD17 pKa = 3.64RR18 pKa = 11.84KK19 pKa = 9.82LPCVNVNIWRR29 pKa = 11.84RR30 pKa = 11.84EE31 pKa = 3.69IEE33 pKa = 4.08YY34 pKa = 10.3KK35 pKa = 11.14ANGVCLNCLNEE46 pKa = 4.79CKK48 pKa = 10.41VCPCDD53 pKa = 3.46YY54 pKa = 10.75CGIRR58 pKa = 11.84HH59 pKa = 5.94KK60 pKa = 11.09CEE62 pKa = 3.44NCLNSDD68 pKa = 4.11CFMNTDD74 pKa = 3.49NEE76 pKa = 4.62FNSHH80 pKa = 5.22RR81 pKa = 11.84WITFDD86 pKa = 4.02EE87 pKa = 5.0EE88 pKa = 4.11PSQMVLFEE96 pKa = 3.83YY97 pKa = 9.86WIMYY101 pKa = 9.91KK102 pKa = 10.47DD103 pKa = 3.64YY104 pKa = 10.65FLSKK108 pKa = 10.48FNYY111 pKa = 9.45NYY113 pKa = 10.52KK114 pKa = 9.94IQLKK118 pKa = 9.07ILNMNKK124 pKa = 9.16NRR126 pKa = 11.84RR127 pKa = 11.84FHH129 pKa = 7.83INEE132 pKa = 4.12SKK134 pKa = 10.67KK135 pKa = 10.41KK136 pKa = 9.48ALSVPITSQYY146 pKa = 11.63LKK148 pKa = 10.79FKK150 pKa = 10.23FNNKK154 pKa = 8.34IYY156 pKa = 10.62IMFGTFLTSKK166 pKa = 8.75IQPWIQLKK174 pKa = 8.78SLKK177 pKa = 10.28VGYY180 pKa = 9.33IQSLNVDD187 pKa = 3.8RR188 pKa = 11.84CAKK191 pKa = 10.54LIATKK196 pKa = 10.81GMFATNSFKK205 pKa = 10.85SSCITEE211 pKa = 3.76INARR215 pKa = 11.84RR216 pKa = 11.84PISEE220 pKa = 4.25CDD222 pKa = 3.59YY223 pKa = 11.23LIEE226 pKa = 5.28ACLCNEE232 pKa = 3.59NNEE235 pKa = 4.35WKK237 pKa = 10.42FSAVMGRR244 pKa = 11.84DD245 pKa = 3.68KK246 pKa = 11.23IPVTKK251 pKa = 10.18SLAMKK256 pKa = 7.77YY257 pKa = 9.59FCKK260 pKa = 10.66NINTEE265 pKa = 3.87LFYY268 pKa = 11.19YY269 pKa = 10.18GHH271 pKa = 7.19AKK273 pKa = 9.52CHH275 pKa = 5.44VVSEE279 pKa = 4.47CPRR282 pKa = 11.84WNQQLRR288 pKa = 11.84VLHH291 pKa = 6.91ASTLNIIFRR300 pKa = 11.84RR301 pKa = 11.84QFMNEE306 pKa = 3.46VVEE309 pKa = 4.18WFEE312 pKa = 5.58NFTQLTGMHH321 pKa = 6.96HH322 pKa = 7.37DD323 pKa = 4.76FIKK326 pKa = 9.28TCVYY330 pKa = 9.97NKK332 pKa = 10.13VIVSHH337 pKa = 5.99FRR339 pKa = 11.84KK340 pKa = 10.18EE341 pKa = 3.63IEE343 pKa = 4.27DD344 pKa = 3.84YY345 pKa = 11.32INFGKK350 pKa = 10.36KK351 pKa = 9.23ISLSSVIPDD360 pKa = 2.95GHH362 pKa = 7.46ALYY365 pKa = 10.95NSIDD369 pKa = 3.66RR370 pKa = 11.84LRR372 pKa = 11.84ISLMLAIDD380 pKa = 3.56VALNRR385 pKa = 11.84IEE387 pKa = 4.11SQQMDD392 pKa = 3.91VLL394 pKa = 4.2
Molecular weight: 46.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5608 |
150 |
1090 |
509.8 |
58.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.456 ± 0.283 | 1.373 ± 0.339 |
5.76 ± 0.276 | 5.617 ± 0.375 |
4.44 ± 0.225 | 3.762 ± 0.322 |
1.819 ± 0.219 | 8.862 ± 0.355 |
6.669 ± 0.588 | 8.06 ± 0.359 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.103 ± 0.173 | 7.4 ± 0.373 |
2.853 ± 0.3 | 3.424 ± 0.374 |
4.547 ± 0.201 | 7.971 ± 0.481 |
6.669 ± 0.387 | 5.831 ± 0.308 |
1.088 ± 0.168 | 5.296 ± 0.494 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
