
Pigeonpea sterility mosaic emaravirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Fimoviridae; Emaravirus
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
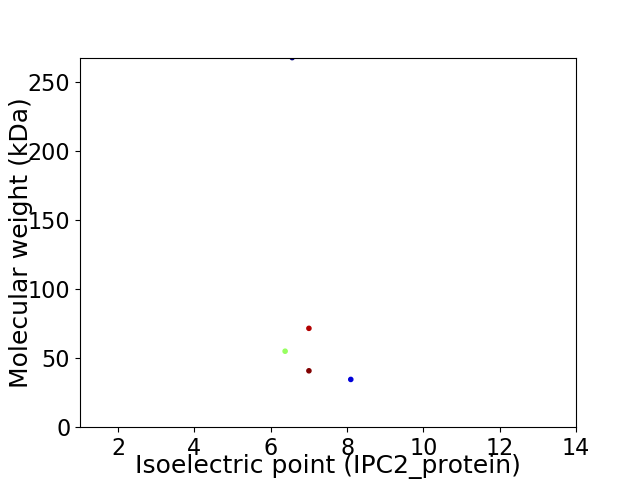
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U6E7B1|U6E7B1_9VIRU p5 OS=Pigeonpea sterility mosaic emaravirus 1 OX=1980429 GN=p5 PE=4 SV=2
MM1 pKa = 7.36EE2 pKa = 6.02SIVPFCISKK11 pKa = 10.71KK12 pKa = 9.11EE13 pKa = 4.01VINNSNYY20 pKa = 10.12EE21 pKa = 3.87KK22 pKa = 10.91LKK24 pKa = 9.91PNKK27 pKa = 9.71NLGRR31 pKa = 11.84MEE33 pKa = 4.72RR34 pKa = 11.84IIYY37 pKa = 10.52DD38 pKa = 3.82MIKK41 pKa = 10.71DD42 pKa = 3.51EE43 pKa = 4.39YY44 pKa = 10.06NRR46 pKa = 11.84CNAVEE51 pKa = 4.7KK52 pKa = 11.13GMFCDD57 pKa = 4.61DD58 pKa = 3.84FNMIPINSCIGYY70 pKa = 8.13MEE72 pKa = 5.28IGDD75 pKa = 5.26DD76 pKa = 3.42IPTYY80 pKa = 10.94VYY82 pKa = 10.52AIKK85 pKa = 10.44KK86 pKa = 8.29GAYY89 pKa = 9.11FKK91 pKa = 11.66GEE93 pKa = 3.63FQYY96 pKa = 11.64AVIEE100 pKa = 4.2LSDD103 pKa = 3.82AVSGAINSILYY114 pKa = 10.01SAVIRR119 pKa = 11.84DD120 pKa = 3.88FEE122 pKa = 6.35GDD124 pKa = 3.33MDD126 pKa = 4.25TLYY129 pKa = 9.5ITVKK133 pKa = 10.18GSLPIKK139 pKa = 9.89EE140 pKa = 3.95YY141 pKa = 10.76FIIVKK146 pKa = 9.89IEE148 pKa = 3.81NKK150 pKa = 7.93QTLIQRR156 pKa = 11.84AKK158 pKa = 10.83SEE160 pKa = 3.9WANILSSTSLVGTDD174 pKa = 4.36SIVSHH179 pKa = 5.41YY180 pKa = 11.13RR181 pKa = 11.84EE182 pKa = 4.01MTRR185 pKa = 11.84FCYY188 pKa = 10.1RR189 pKa = 11.84IIKK192 pKa = 8.65NQSDD196 pKa = 3.76VGSVYY201 pKa = 11.47SMISEE206 pKa = 4.03NQHH209 pKa = 5.66LVNHH213 pKa = 6.19VLPSLTMYY221 pKa = 8.11LTSYY225 pKa = 10.78NSVILEE231 pKa = 4.05HH232 pKa = 6.44VKK234 pKa = 10.48NHH236 pKa = 5.99IKK238 pKa = 10.25RR239 pKa = 11.84CYY241 pKa = 9.85EE242 pKa = 3.95SNDD245 pKa = 3.15YY246 pKa = 11.37NLIFRR251 pKa = 11.84KK252 pKa = 10.11DD253 pKa = 3.93DD254 pKa = 3.53IEE256 pKa = 5.58CDD258 pKa = 3.63HH259 pKa = 6.47LTEE262 pKa = 4.75PYY264 pKa = 9.75IDD266 pKa = 4.1EE267 pKa = 4.32GLKK270 pKa = 10.87LEE272 pKa = 4.46ISNAIYY278 pKa = 10.97NDD280 pKa = 2.9VTLRR284 pKa = 11.84LRR286 pKa = 11.84EE287 pKa = 4.07IKK289 pKa = 10.59NCEE292 pKa = 3.82AEE294 pKa = 3.84AKK296 pKa = 10.03YY297 pKa = 10.3IKK299 pKa = 10.49KK300 pKa = 10.24NLFRR304 pKa = 11.84SDD306 pKa = 3.32NNKK309 pKa = 9.98IISDD313 pKa = 4.01FEE315 pKa = 4.33AVTSSEE321 pKa = 4.14TEE323 pKa = 3.9MIKK326 pKa = 10.41YY327 pKa = 10.52LPGTNEE333 pKa = 3.87AEE335 pKa = 5.28DD336 pKa = 4.01FFQYY340 pKa = 11.06GLPEE344 pKa = 5.02HH345 pKa = 6.85INIARR350 pKa = 11.84THH352 pKa = 6.74LEE354 pKa = 3.3NSMVKK359 pKa = 10.39RR360 pKa = 11.84ITDD363 pKa = 3.21ANMIDD368 pKa = 3.95DD369 pKa = 3.79VDD371 pKa = 4.62YY372 pKa = 11.0IPSYY376 pKa = 11.01PFTTEE381 pKa = 3.49FKK383 pKa = 10.35KK384 pKa = 11.03VVFEE388 pKa = 4.3SYY390 pKa = 10.17RR391 pKa = 11.84IKK393 pKa = 11.12NKK395 pKa = 6.63TVKK398 pKa = 9.43MSHH401 pKa = 6.72LGLHH405 pKa = 5.6YY406 pKa = 10.59ACLYY410 pKa = 9.82IYY412 pKa = 9.96FSIVSTRR419 pKa = 11.84RR420 pKa = 11.84AGYY423 pKa = 9.53EE424 pKa = 3.76RR425 pKa = 11.84FNFDD429 pKa = 2.97HH430 pKa = 7.07KK431 pKa = 11.07LIEE434 pKa = 4.21KK435 pKa = 9.74SRR437 pKa = 11.84KK438 pKa = 6.85FAKK441 pKa = 10.6VNIEE445 pKa = 3.72YY446 pKa = 10.69LRR448 pKa = 11.84FRR450 pKa = 11.84IQTVRR455 pKa = 11.84TVSTPINNCVTITLKK470 pKa = 11.04KK471 pKa = 10.49NLL473 pKa = 3.98
MM1 pKa = 7.36EE2 pKa = 6.02SIVPFCISKK11 pKa = 10.71KK12 pKa = 9.11EE13 pKa = 4.01VINNSNYY20 pKa = 10.12EE21 pKa = 3.87KK22 pKa = 10.91LKK24 pKa = 9.91PNKK27 pKa = 9.71NLGRR31 pKa = 11.84MEE33 pKa = 4.72RR34 pKa = 11.84IIYY37 pKa = 10.52DD38 pKa = 3.82MIKK41 pKa = 10.71DD42 pKa = 3.51EE43 pKa = 4.39YY44 pKa = 10.06NRR46 pKa = 11.84CNAVEE51 pKa = 4.7KK52 pKa = 11.13GMFCDD57 pKa = 4.61DD58 pKa = 3.84FNMIPINSCIGYY70 pKa = 8.13MEE72 pKa = 5.28IGDD75 pKa = 5.26DD76 pKa = 3.42IPTYY80 pKa = 10.94VYY82 pKa = 10.52AIKK85 pKa = 10.44KK86 pKa = 8.29GAYY89 pKa = 9.11FKK91 pKa = 11.66GEE93 pKa = 3.63FQYY96 pKa = 11.64AVIEE100 pKa = 4.2LSDD103 pKa = 3.82AVSGAINSILYY114 pKa = 10.01SAVIRR119 pKa = 11.84DD120 pKa = 3.88FEE122 pKa = 6.35GDD124 pKa = 3.33MDD126 pKa = 4.25TLYY129 pKa = 9.5ITVKK133 pKa = 10.18GSLPIKK139 pKa = 9.89EE140 pKa = 3.95YY141 pKa = 10.76FIIVKK146 pKa = 9.89IEE148 pKa = 3.81NKK150 pKa = 7.93QTLIQRR156 pKa = 11.84AKK158 pKa = 10.83SEE160 pKa = 3.9WANILSSTSLVGTDD174 pKa = 4.36SIVSHH179 pKa = 5.41YY180 pKa = 11.13RR181 pKa = 11.84EE182 pKa = 4.01MTRR185 pKa = 11.84FCYY188 pKa = 10.1RR189 pKa = 11.84IIKK192 pKa = 8.65NQSDD196 pKa = 3.76VGSVYY201 pKa = 11.47SMISEE206 pKa = 4.03NQHH209 pKa = 5.66LVNHH213 pKa = 6.19VLPSLTMYY221 pKa = 8.11LTSYY225 pKa = 10.78NSVILEE231 pKa = 4.05HH232 pKa = 6.44VKK234 pKa = 10.48NHH236 pKa = 5.99IKK238 pKa = 10.25RR239 pKa = 11.84CYY241 pKa = 9.85EE242 pKa = 3.95SNDD245 pKa = 3.15YY246 pKa = 11.37NLIFRR251 pKa = 11.84KK252 pKa = 10.11DD253 pKa = 3.93DD254 pKa = 3.53IEE256 pKa = 5.58CDD258 pKa = 3.63HH259 pKa = 6.47LTEE262 pKa = 4.75PYY264 pKa = 9.75IDD266 pKa = 4.1EE267 pKa = 4.32GLKK270 pKa = 10.87LEE272 pKa = 4.46ISNAIYY278 pKa = 10.97NDD280 pKa = 2.9VTLRR284 pKa = 11.84LRR286 pKa = 11.84EE287 pKa = 4.07IKK289 pKa = 10.59NCEE292 pKa = 3.82AEE294 pKa = 3.84AKK296 pKa = 10.03YY297 pKa = 10.3IKK299 pKa = 10.49KK300 pKa = 10.24NLFRR304 pKa = 11.84SDD306 pKa = 3.32NNKK309 pKa = 9.98IISDD313 pKa = 4.01FEE315 pKa = 4.33AVTSSEE321 pKa = 4.14TEE323 pKa = 3.9MIKK326 pKa = 10.41YY327 pKa = 10.52LPGTNEE333 pKa = 3.87AEE335 pKa = 5.28DD336 pKa = 4.01FFQYY340 pKa = 11.06GLPEE344 pKa = 5.02HH345 pKa = 6.85INIARR350 pKa = 11.84THH352 pKa = 6.74LEE354 pKa = 3.3NSMVKK359 pKa = 10.39RR360 pKa = 11.84ITDD363 pKa = 3.21ANMIDD368 pKa = 3.95DD369 pKa = 3.79VDD371 pKa = 4.62YY372 pKa = 11.0IPSYY376 pKa = 11.01PFTTEE381 pKa = 3.49FKK383 pKa = 10.35KK384 pKa = 11.03VVFEE388 pKa = 4.3SYY390 pKa = 10.17RR391 pKa = 11.84IKK393 pKa = 11.12NKK395 pKa = 6.63TVKK398 pKa = 9.43MSHH401 pKa = 6.72LGLHH405 pKa = 5.6YY406 pKa = 10.59ACLYY410 pKa = 9.82IYY412 pKa = 9.96FSIVSTRR419 pKa = 11.84RR420 pKa = 11.84AGYY423 pKa = 9.53EE424 pKa = 3.76RR425 pKa = 11.84FNFDD429 pKa = 2.97HH430 pKa = 7.07KK431 pKa = 11.07LIEE434 pKa = 4.21KK435 pKa = 9.74SRR437 pKa = 11.84KK438 pKa = 6.85FAKK441 pKa = 10.6VNIEE445 pKa = 3.72YY446 pKa = 10.69LRR448 pKa = 11.84FRR450 pKa = 11.84IQTVRR455 pKa = 11.84TVSTPINNCVTITLKK470 pKa = 11.04KK471 pKa = 10.49NLL473 pKa = 3.98
Molecular weight: 55.08 kDa
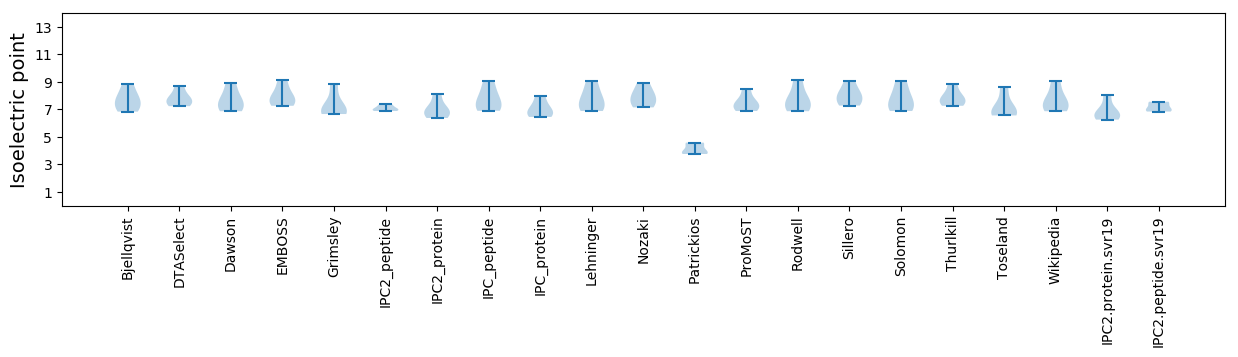
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U6CPM6|U6CPM6_9VIRU Replicase OS=Pigeonpea sterility mosaic emaravirus 1 OX=1980429 GN=RdRp PE=4 SV=1
MM1 pKa = 7.36PPKK4 pKa = 9.85MPSKK8 pKa = 9.75TPLSNMPAASKK19 pKa = 10.81KK20 pKa = 10.57SDD22 pKa = 3.23IPDD25 pKa = 3.27NEE27 pKa = 3.46VRR29 pKa = 11.84INVKK33 pKa = 10.61GKK35 pKa = 9.9VNTIKK40 pKa = 10.64FPNVKK45 pKa = 9.92DD46 pKa = 3.58KK47 pKa = 11.12KK48 pKa = 9.75LQNTEE53 pKa = 3.99GTSLKK58 pKa = 10.23PATIEE63 pKa = 3.78PKK65 pKa = 10.26AFKK68 pKa = 10.06SAAMVEE74 pKa = 4.08HH75 pKa = 7.18DD76 pKa = 4.75FKK78 pKa = 10.8IKK80 pKa = 10.45KK81 pKa = 9.39YY82 pKa = 10.57ISYY85 pKa = 10.85CNINAAAAYY94 pKa = 9.44LSQSKK99 pKa = 7.37EE100 pKa = 3.86HH101 pKa = 7.78KK102 pKa = 10.54EE103 pKa = 3.56MLKK106 pKa = 10.74QNDD109 pKa = 3.97SLILTVSKK117 pKa = 10.38DD118 pKa = 3.14QKK120 pKa = 10.71IYY122 pKa = 10.7VIQNIQEE129 pKa = 4.23TNIEE133 pKa = 4.1NVLSFNKK140 pKa = 10.08ACAVLALGILKK151 pKa = 10.36HH152 pKa = 6.14KK153 pKa = 10.27FPEE156 pKa = 4.25IFDD159 pKa = 4.09WTSHH163 pKa = 6.16KK164 pKa = 10.75YY165 pKa = 7.65VTSGWSDD172 pKa = 3.2QNLNVEE178 pKa = 4.09DD179 pKa = 4.39TIINRR184 pKa = 11.84LAGAMGLTPDD194 pKa = 3.44NPYY197 pKa = 10.23YY198 pKa = 10.2WFMVPGYY205 pKa = 9.16EE206 pKa = 4.82FLYY209 pKa = 10.17EE210 pKa = 4.63LYY212 pKa = 9.83PAEE215 pKa = 4.83CIAYY219 pKa = 7.55TLLRR223 pKa = 11.84VEE225 pKa = 4.11YY226 pKa = 10.34RR227 pKa = 11.84EE228 pKa = 3.9VLNIPEE234 pKa = 5.0KK235 pKa = 10.56ISNQDD240 pKa = 2.64IVQSLTAKK248 pKa = 9.15MNKK251 pKa = 8.87FHH253 pKa = 6.95GLEE256 pKa = 3.84TSTFKK261 pKa = 11.11DD262 pKa = 3.21AVAVVGLEE270 pKa = 4.1NIKK273 pKa = 10.42AAYY276 pKa = 9.19QAMSSSVGEE285 pKa = 3.97TGRR288 pKa = 11.84TRR290 pKa = 11.84RR291 pKa = 11.84AAIVLAGFEE300 pKa = 4.38EE301 pKa = 4.5LLKK304 pKa = 10.79SLKK307 pKa = 10.1EE308 pKa = 3.89
MM1 pKa = 7.36PPKK4 pKa = 9.85MPSKK8 pKa = 9.75TPLSNMPAASKK19 pKa = 10.81KK20 pKa = 10.57SDD22 pKa = 3.23IPDD25 pKa = 3.27NEE27 pKa = 3.46VRR29 pKa = 11.84INVKK33 pKa = 10.61GKK35 pKa = 9.9VNTIKK40 pKa = 10.64FPNVKK45 pKa = 9.92DD46 pKa = 3.58KK47 pKa = 11.12KK48 pKa = 9.75LQNTEE53 pKa = 3.99GTSLKK58 pKa = 10.23PATIEE63 pKa = 3.78PKK65 pKa = 10.26AFKK68 pKa = 10.06SAAMVEE74 pKa = 4.08HH75 pKa = 7.18DD76 pKa = 4.75FKK78 pKa = 10.8IKK80 pKa = 10.45KK81 pKa = 9.39YY82 pKa = 10.57ISYY85 pKa = 10.85CNINAAAAYY94 pKa = 9.44LSQSKK99 pKa = 7.37EE100 pKa = 3.86HH101 pKa = 7.78KK102 pKa = 10.54EE103 pKa = 3.56MLKK106 pKa = 10.74QNDD109 pKa = 3.97SLILTVSKK117 pKa = 10.38DD118 pKa = 3.14QKK120 pKa = 10.71IYY122 pKa = 10.7VIQNIQEE129 pKa = 4.23TNIEE133 pKa = 4.1NVLSFNKK140 pKa = 10.08ACAVLALGILKK151 pKa = 10.36HH152 pKa = 6.14KK153 pKa = 10.27FPEE156 pKa = 4.25IFDD159 pKa = 4.09WTSHH163 pKa = 6.16KK164 pKa = 10.75YY165 pKa = 7.65VTSGWSDD172 pKa = 3.2QNLNVEE178 pKa = 4.09DD179 pKa = 4.39TIINRR184 pKa = 11.84LAGAMGLTPDD194 pKa = 3.44NPYY197 pKa = 10.23YY198 pKa = 10.2WFMVPGYY205 pKa = 9.16EE206 pKa = 4.82FLYY209 pKa = 10.17EE210 pKa = 4.63LYY212 pKa = 9.83PAEE215 pKa = 4.83CIAYY219 pKa = 7.55TLLRR223 pKa = 11.84VEE225 pKa = 4.11YY226 pKa = 10.34RR227 pKa = 11.84EE228 pKa = 3.9VLNIPEE234 pKa = 5.0KK235 pKa = 10.56ISNQDD240 pKa = 2.64IVQSLTAKK248 pKa = 9.15MNKK251 pKa = 8.87FHH253 pKa = 6.95GLEE256 pKa = 3.84TSTFKK261 pKa = 11.11DD262 pKa = 3.21AVAVVGLEE270 pKa = 4.1NIKK273 pKa = 10.42AAYY276 pKa = 9.19QAMSSSVGEE285 pKa = 3.97TGRR288 pKa = 11.84TRR290 pKa = 11.84RR291 pKa = 11.84AAIVLAGFEE300 pKa = 4.38EE301 pKa = 4.5LLKK304 pKa = 10.79SLKK307 pKa = 10.1EE308 pKa = 3.89
Molecular weight: 34.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4059 |
308 |
2294 |
811.8 |
94.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.523 ± 0.749 | 1.897 ± 0.436 |
6.332 ± 0.365 | 5.79 ± 0.415 |
4.484 ± 0.272 | 3.646 ± 0.695 |
2.488 ± 0.234 | 8.943 ± 0.632 |
8.426 ± 0.368 | 8.795 ± 0.906 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.956 ± 0.145 | 6.948 ± 0.65 |
3.277 ± 0.262 | 2.685 ± 0.328 |
3.819 ± 0.304 | 7.982 ± 0.378 |
6.011 ± 0.349 | 5.494 ± 0.228 |
0.616 ± 0.14 | 5.888 ± 0.503 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
