
Loktanella sp. S4079
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Loktanella; unclassified Loktanella
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
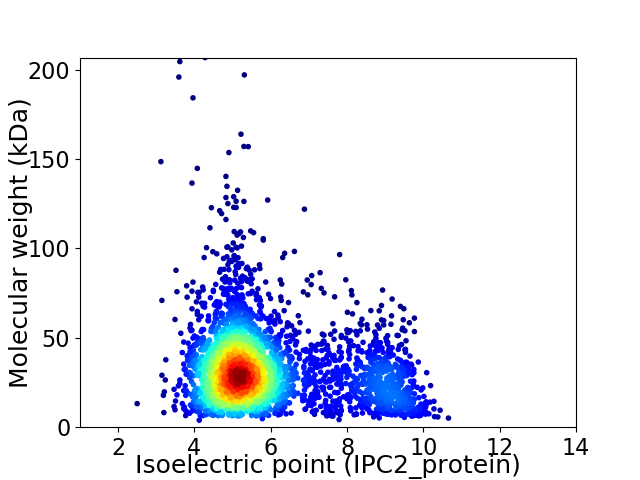
Virtual 2D-PAGE plot for 3296 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F4RL21|A0A0F4RL21_9RHOB Chemotaxis protein CheY OS=Loktanella sp. S4079 OX=579483 GN=TW80_11820 PE=4 SV=1
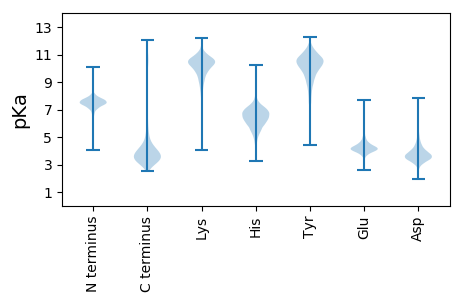
MM1 pKa = 7.33LRR3 pKa = 11.84WWVNADD9 pKa = 3.48RR10 pKa = 11.84KK11 pKa = 8.69VTIVTITQKK20 pKa = 9.93TIFPKK25 pKa = 10.61RR26 pKa = 11.84FDD28 pKa = 3.55RR29 pKa = 11.84KK30 pKa = 10.42AIAVLAALSLSSALVTPDD48 pKa = 3.77RR49 pKa = 11.84LDD51 pKa = 4.05AGDD54 pKa = 4.68FTFDD58 pKa = 3.09WSVIDD63 pKa = 3.73WPEE66 pKa = 3.7GSLGPNTYY74 pKa = 8.71TLTDD78 pKa = 3.23QYY80 pKa = 11.59GFQIDD85 pKa = 3.74SRR87 pKa = 11.84IQMSGIFEE95 pKa = 4.65SYY97 pKa = 11.04GPANSPNDD105 pKa = 3.58DD106 pKa = 3.85VIFGGNVEE114 pKa = 4.22SLILVSDD121 pKa = 4.21APVGSANVGDD131 pKa = 4.08ATVSTTLSFSSGGIVFPVDD150 pKa = 3.27GLEE153 pKa = 4.83LRR155 pKa = 11.84VLDD158 pKa = 5.72IDD160 pKa = 4.12ATDD163 pKa = 3.84NNNASDD169 pKa = 3.5RR170 pKa = 11.84CDD172 pKa = 3.32FVTLTGNNGNPLLTVASATPTVLVGPGPGSGGTGALAANQAQCIYY217 pKa = 10.56VDD219 pKa = 3.96GFGASPTSNNDD230 pKa = 3.08DD231 pKa = 3.64TGTIIATYY239 pKa = 10.27PNDD242 pKa = 3.77TSSTTIFYY250 pKa = 10.66DD251 pKa = 3.58EE252 pKa = 4.96SIGNVRR258 pKa = 11.84PLFLGEE264 pKa = 4.32DD265 pKa = 3.51PAARR269 pKa = 11.84GIGVLGDD276 pKa = 3.07IAFNVDD282 pKa = 3.13QSISLSRR289 pKa = 11.84SVTPTTGLAGDD300 pKa = 4.07SVTYY304 pKa = 10.12EE305 pKa = 3.67YY306 pKa = 11.49VVTNNGALPFNTGQDD321 pKa = 4.05LIIEE325 pKa = 4.81DD326 pKa = 4.66DD327 pKa = 4.13LLGTVTCPAITAPVAPGGTVTCTAPYY353 pKa = 9.78TITPADD359 pKa = 3.81ALTGSVNSNATAGIGAIGQPFATRR383 pKa = 11.84LQSNTANLNVVTITPPGEE401 pKa = 4.49PGPQTCSPEE410 pKa = 4.35SVFSKK415 pKa = 9.73TRR417 pKa = 11.84TQLAGPGSAAAPTTSDD433 pKa = 2.4IFLYY437 pKa = 10.88DD438 pKa = 4.3DD439 pKa = 3.57VTFDD443 pKa = 4.0TNGNPIDD450 pKa = 4.32IVMQLQEE457 pKa = 4.14ISNAINVEE465 pKa = 3.81ISSGLEE471 pKa = 3.58ARR473 pKa = 11.84MVPADD478 pKa = 3.85NGYY481 pKa = 8.17VTYY484 pKa = 10.67SLRR487 pKa = 11.84LVQDD491 pKa = 4.19GSATPSNPQGTPIEE505 pKa = 4.05LSRR508 pKa = 11.84INGVIVQQTDD518 pKa = 2.64VDD520 pKa = 4.03SRR522 pKa = 11.84GAGDD526 pKa = 4.18DD527 pKa = 3.56SSDD530 pKa = 3.68VVGVSLTPTTLTHH543 pKa = 7.21FNTAPLASFPAAGTAIAMDD562 pKa = 4.17PAKK565 pKa = 10.34TGDD568 pKa = 3.53PTNWTDD574 pKa = 4.04EE575 pKa = 4.37PNEE578 pKa = 4.16TAFDD582 pKa = 3.69NYY584 pKa = 8.13VTYY587 pKa = 10.61EE588 pKa = 3.8FDD590 pKa = 3.46SFQEE594 pKa = 4.26GTFIHH599 pKa = 7.43GYY601 pKa = 7.93TGTSINPATRR611 pKa = 11.84GSGILLCAIANTSPTVVAEE630 pKa = 4.42DD631 pKa = 4.31DD632 pKa = 5.23DD633 pKa = 4.53YY634 pKa = 11.79TATPINSVGGGTTGEE649 pKa = 4.08VMANDD654 pKa = 5.05TINGLPATFLNATISVISEE673 pKa = 4.0ATPVTTGALVPTLVTSGVDD692 pKa = 3.12EE693 pKa = 4.59GRR695 pKa = 11.84VVVPDD700 pKa = 3.92GVPAGNYY707 pKa = 9.05FIEE710 pKa = 4.42YY711 pKa = 9.27LLCDD715 pKa = 3.5ATDD718 pKa = 3.46TTDD721 pKa = 3.34CDD723 pKa = 3.61RR724 pKa = 11.84AKK726 pKa = 10.14VTISVYY732 pKa = 10.64DD733 pKa = 3.51GNGFDD738 pKa = 5.84FGDD741 pKa = 4.35APPSYY746 pKa = 9.23LTASHH751 pKa = 6.73GVPDD755 pKa = 3.98TPVIYY760 pKa = 10.48LGVVPPDD767 pKa = 3.61AEE769 pKa = 4.77LVAQSDD775 pKa = 3.93ATATADD781 pKa = 4.17DD782 pKa = 4.4VFNIDD787 pKa = 5.34DD788 pKa = 3.98EE789 pKa = 4.91DD790 pKa = 4.0AVSFPILTQGTISTLDD806 pKa = 3.17VGVTGNGTLQAWIDD820 pKa = 3.87FNGDD824 pKa = 3.3GLFEE828 pKa = 4.05QTLGEE833 pKa = 4.8RR834 pKa = 11.84IATDD838 pKa = 3.51LRR840 pKa = 11.84DD841 pKa = 4.26DD842 pKa = 4.36GSAFDD847 pKa = 4.63NVAGDD852 pKa = 3.85GVIQIDD858 pKa = 3.87VAVPTDD864 pKa = 3.4ATTDD868 pKa = 3.2TTFARR873 pKa = 11.84FRR875 pKa = 11.84YY876 pKa = 7.75STSPGLSPTDD886 pKa = 3.77FAPDD890 pKa = 3.92GEE892 pKa = 4.53VEE894 pKa = 4.61DD895 pKa = 4.39YY896 pKa = 11.45SLVIAAADD904 pKa = 3.6LVDD907 pKa = 5.34RR908 pKa = 11.84GDD910 pKa = 3.89APASYY915 pKa = 10.6GDD917 pKa = 3.68PRR919 pKa = 11.84HH920 pKa = 6.61IIVDD924 pKa = 4.47DD925 pKa = 3.73IFLGAGIPDD934 pKa = 4.54SEE936 pKa = 4.42TDD938 pKa = 3.5PQNSVDD944 pKa = 4.55ADD946 pKa = 3.73ADD948 pKa = 3.99DD949 pKa = 4.19FAGADD954 pKa = 4.15DD955 pKa = 4.43EE956 pKa = 5.92DD957 pKa = 4.53SVAVFPVFEE966 pKa = 4.35VGTTVSVTVQTHH978 pKa = 4.6EE979 pKa = 4.31TLSLQYY985 pKa = 11.49ALGIPVTSGVTNLQAWIDD1003 pKa = 3.7FDD1005 pKa = 4.34QNGTFEE1011 pKa = 4.48PSEE1014 pKa = 4.02QVATNYY1020 pKa = 10.17RR1021 pKa = 11.84DD1022 pKa = 3.49GGIGDD1027 pKa = 3.69TDD1029 pKa = 4.73GIFNNQISFDD1039 pKa = 3.68VTVPNTISSGYY1050 pKa = 8.49TYY1052 pKa = 11.42ARR1054 pKa = 11.84LRR1056 pKa = 11.84WSTSSALVSDD1066 pKa = 4.82PFDD1069 pKa = 3.53GLNFDD1074 pKa = 5.32GEE1076 pKa = 4.54VEE1078 pKa = 4.22DD1079 pKa = 4.61YY1080 pKa = 11.11EE1081 pKa = 4.75VVLSAGAVPFEE1092 pKa = 4.74CDD1094 pKa = 2.74GTLYY1098 pKa = 10.33RR1099 pKa = 11.84VARR1102 pKa = 11.84VDD1104 pKa = 3.36SQLQRR1109 pKa = 11.84LVFSEE1114 pKa = 4.78DD1115 pKa = 2.93GSGGYY1120 pKa = 8.7TIDD1123 pKa = 3.62ATNVGAPAGVAYY1135 pKa = 10.79NGGWGYY1141 pKa = 10.77NAVDD1145 pKa = 3.37GLFYY1149 pKa = 10.69GVRR1152 pKa = 11.84EE1153 pKa = 4.1FEE1155 pKa = 4.22RR1156 pKa = 11.84DD1157 pKa = 3.38LVQLDD1162 pKa = 3.61STGKK1166 pKa = 8.01FTVVSTLPPTAATGYY1181 pKa = 10.28NSGDD1185 pKa = 3.33ILSNGIMIYY1194 pKa = 10.11RR1195 pKa = 11.84VQGTNDD1201 pKa = 3.36FQLLDD1206 pKa = 3.57ITDD1209 pKa = 3.77PTNPIDD1215 pKa = 4.21AGLVTLTSAVDD1226 pKa = 3.57PFDD1229 pKa = 4.03IAFNPNDD1236 pKa = 3.21GMIYY1240 pKa = 10.21GVNHH1244 pKa = 4.57VTDD1247 pKa = 3.56RR1248 pKa = 11.84LFYY1251 pKa = 10.55FDD1253 pKa = 5.09PADD1256 pKa = 3.98GAPGTRR1262 pKa = 11.84TPVEE1266 pKa = 4.27FGPAIWTEE1274 pKa = 3.9AYY1276 pKa = 9.86GAIWFDD1282 pKa = 3.72FYY1284 pKa = 11.75GRR1286 pKa = 11.84MYY1288 pKa = 11.16VNQNTSNEE1296 pKa = 4.14MYY1298 pKa = 9.96EE1299 pKa = 4.04VDD1301 pKa = 5.34VGIQGNGTGNRR1312 pKa = 11.84EE1313 pKa = 4.64LISVITISEE1322 pKa = 4.07EE1323 pKa = 3.7FRR1325 pKa = 11.84NDD1327 pKa = 3.26GAGCPSRR1334 pKa = 11.84LGPLPPEE1341 pKa = 4.25GALAGTVYY1349 pKa = 10.87EE1350 pKa = 4.56DD1351 pKa = 3.9TDD1353 pKa = 3.91GSGAFDD1359 pKa = 3.57NGEE1362 pKa = 3.98TGIPNISVDD1371 pKa = 3.23IYY1373 pKa = 11.2NDD1375 pKa = 2.99NGTPGTIVDD1384 pKa = 4.32DD1385 pKa = 4.08VLVTSVVTAADD1396 pKa = 3.52GSYY1399 pKa = 10.62LVEE1402 pKa = 4.65NLSALNTYY1410 pKa = 10.23RR1411 pKa = 11.84IEE1413 pKa = 4.6VEE1415 pKa = 4.61DD1416 pKa = 4.96ADD1418 pKa = 4.41ADD1420 pKa = 3.99MPAGYY1425 pKa = 10.18QSVTPNPLTAIRR1437 pKa = 11.84VTAGSTRR1444 pKa = 11.84GGLDD1448 pKa = 3.13FGFAPGEE1455 pKa = 4.01ADD1457 pKa = 3.97LSITKK1462 pKa = 8.87TVEE1465 pKa = 3.87RR1466 pKa = 11.84TSDD1469 pKa = 3.29GATVTSAAEE1478 pKa = 4.1GTEE1481 pKa = 4.07LEE1483 pKa = 4.41FVISVTNSGPASVASVLVNDD1503 pKa = 4.68ILPSGFAYY1511 pKa = 10.76VSDD1514 pKa = 4.36DD1515 pKa = 3.92AASQGNTYY1523 pKa = 10.3NASTGRR1529 pKa = 11.84WDD1531 pKa = 3.56VGGLAIGEE1539 pKa = 4.47TKK1541 pKa = 9.38TLRR1544 pKa = 11.84ITVTMLGSGNHH1555 pKa = 4.91TNIAEE1560 pKa = 4.48IIFSSVDD1567 pKa = 3.83DD1568 pKa = 4.76PDD1570 pKa = 4.33SDD1572 pKa = 4.14PSVGTATDD1580 pKa = 4.13DD1581 pKa = 4.14LSDD1584 pKa = 5.47GIPDD1588 pKa = 4.14DD1589 pKa = 5.58DD1590 pKa = 4.07EE1591 pKa = 7.09ASVTVALDD1599 pKa = 3.16NGEE1602 pKa = 4.22RR1603 pKa = 11.84LLSGRR1608 pKa = 11.84VFIDD1612 pKa = 2.9NGVNSGTAHH1621 pKa = 6.74DD1622 pKa = 5.25AIVNGSEE1629 pKa = 4.19TGTQSATLTILDD1641 pKa = 3.89GSGSVIATPTIAADD1655 pKa = 4.13GSWAYY1660 pKa = 10.58ALDD1663 pKa = 3.98GSYY1666 pKa = 10.49SGPLSVSVAPLPGFITVSEE1685 pKa = 4.23NDD1687 pKa = 3.22SGLPALANSDD1697 pKa = 3.45PHH1699 pKa = 7.71DD1700 pKa = 4.32GNFTFTPDD1708 pKa = 2.93ASSNYY1713 pKa = 9.45SGLNIGIVRR1722 pKa = 11.84APTLTRR1728 pKa = 11.84DD1729 pKa = 3.32QEE1731 pKa = 4.22ASVEE1735 pKa = 4.25SGQVATLQHH1744 pKa = 7.28EE1745 pKa = 4.69YY1746 pKa = 9.84TAFTSGTVTFSYY1758 pKa = 10.82ANPNMAPPNSFSATLYY1774 pKa = 10.72RR1775 pKa = 11.84DD1776 pKa = 3.75TDD1778 pKa = 3.92CDD1780 pKa = 3.91TTPDD1784 pKa = 3.66TPINAPITVSAGDD1797 pKa = 3.8RR1798 pKa = 11.84LCLVSRR1804 pKa = 11.84VTANSGIGPGSSYY1817 pKa = 10.78TYY1819 pKa = 11.15DD1820 pKa = 3.52LVASTALSATTVVSTSQNTDD1840 pKa = 3.46RR1841 pKa = 11.84IVAGGGAGRR1850 pKa = 11.84LEE1852 pKa = 3.87LRR1854 pKa = 11.84KK1855 pKa = 7.74TVRR1858 pKa = 11.84NITQNTPEE1866 pKa = 4.26GASNGGAIGDD1876 pKa = 3.65VLRR1879 pKa = 11.84YY1880 pKa = 9.56QIYY1883 pKa = 10.51LRR1885 pKa = 11.84NPSSAPATNVIIYY1898 pKa = 9.45DD1899 pKa = 3.59KK1900 pKa = 9.93TPSYY1904 pKa = 9.52TALASPIASPTTLGNNLICVVSQPTNNVAGYY1935 pKa = 9.46VGPLRR1940 pKa = 11.84WDD1942 pKa = 3.32CTGTYY1947 pKa = 10.35EE1948 pKa = 4.92PGEE1951 pKa = 4.45DD1952 pKa = 3.26DD1953 pKa = 4.73SVFFDD1958 pKa = 4.36VVISPP1963 pKa = 4.35
MM1 pKa = 7.33LRR3 pKa = 11.84WWVNADD9 pKa = 3.48RR10 pKa = 11.84KK11 pKa = 8.69VTIVTITQKK20 pKa = 9.93TIFPKK25 pKa = 10.61RR26 pKa = 11.84FDD28 pKa = 3.55RR29 pKa = 11.84KK30 pKa = 10.42AIAVLAALSLSSALVTPDD48 pKa = 3.77RR49 pKa = 11.84LDD51 pKa = 4.05AGDD54 pKa = 4.68FTFDD58 pKa = 3.09WSVIDD63 pKa = 3.73WPEE66 pKa = 3.7GSLGPNTYY74 pKa = 8.71TLTDD78 pKa = 3.23QYY80 pKa = 11.59GFQIDD85 pKa = 3.74SRR87 pKa = 11.84IQMSGIFEE95 pKa = 4.65SYY97 pKa = 11.04GPANSPNDD105 pKa = 3.58DD106 pKa = 3.85VIFGGNVEE114 pKa = 4.22SLILVSDD121 pKa = 4.21APVGSANVGDD131 pKa = 4.08ATVSTTLSFSSGGIVFPVDD150 pKa = 3.27GLEE153 pKa = 4.83LRR155 pKa = 11.84VLDD158 pKa = 5.72IDD160 pKa = 4.12ATDD163 pKa = 3.84NNNASDD169 pKa = 3.5RR170 pKa = 11.84CDD172 pKa = 3.32FVTLTGNNGNPLLTVASATPTVLVGPGPGSGGTGALAANQAQCIYY217 pKa = 10.56VDD219 pKa = 3.96GFGASPTSNNDD230 pKa = 3.08DD231 pKa = 3.64TGTIIATYY239 pKa = 10.27PNDD242 pKa = 3.77TSSTTIFYY250 pKa = 10.66DD251 pKa = 3.58EE252 pKa = 4.96SIGNVRR258 pKa = 11.84PLFLGEE264 pKa = 4.32DD265 pKa = 3.51PAARR269 pKa = 11.84GIGVLGDD276 pKa = 3.07IAFNVDD282 pKa = 3.13QSISLSRR289 pKa = 11.84SVTPTTGLAGDD300 pKa = 4.07SVTYY304 pKa = 10.12EE305 pKa = 3.67YY306 pKa = 11.49VVTNNGALPFNTGQDD321 pKa = 4.05LIIEE325 pKa = 4.81DD326 pKa = 4.66DD327 pKa = 4.13LLGTVTCPAITAPVAPGGTVTCTAPYY353 pKa = 9.78TITPADD359 pKa = 3.81ALTGSVNSNATAGIGAIGQPFATRR383 pKa = 11.84LQSNTANLNVVTITPPGEE401 pKa = 4.49PGPQTCSPEE410 pKa = 4.35SVFSKK415 pKa = 9.73TRR417 pKa = 11.84TQLAGPGSAAAPTTSDD433 pKa = 2.4IFLYY437 pKa = 10.88DD438 pKa = 4.3DD439 pKa = 3.57VTFDD443 pKa = 4.0TNGNPIDD450 pKa = 4.32IVMQLQEE457 pKa = 4.14ISNAINVEE465 pKa = 3.81ISSGLEE471 pKa = 3.58ARR473 pKa = 11.84MVPADD478 pKa = 3.85NGYY481 pKa = 8.17VTYY484 pKa = 10.67SLRR487 pKa = 11.84LVQDD491 pKa = 4.19GSATPSNPQGTPIEE505 pKa = 4.05LSRR508 pKa = 11.84INGVIVQQTDD518 pKa = 2.64VDD520 pKa = 4.03SRR522 pKa = 11.84GAGDD526 pKa = 4.18DD527 pKa = 3.56SSDD530 pKa = 3.68VVGVSLTPTTLTHH543 pKa = 7.21FNTAPLASFPAAGTAIAMDD562 pKa = 4.17PAKK565 pKa = 10.34TGDD568 pKa = 3.53PTNWTDD574 pKa = 4.04EE575 pKa = 4.37PNEE578 pKa = 4.16TAFDD582 pKa = 3.69NYY584 pKa = 8.13VTYY587 pKa = 10.61EE588 pKa = 3.8FDD590 pKa = 3.46SFQEE594 pKa = 4.26GTFIHH599 pKa = 7.43GYY601 pKa = 7.93TGTSINPATRR611 pKa = 11.84GSGILLCAIANTSPTVVAEE630 pKa = 4.42DD631 pKa = 4.31DD632 pKa = 5.23DD633 pKa = 4.53YY634 pKa = 11.79TATPINSVGGGTTGEE649 pKa = 4.08VMANDD654 pKa = 5.05TINGLPATFLNATISVISEE673 pKa = 4.0ATPVTTGALVPTLVTSGVDD692 pKa = 3.12EE693 pKa = 4.59GRR695 pKa = 11.84VVVPDD700 pKa = 3.92GVPAGNYY707 pKa = 9.05FIEE710 pKa = 4.42YY711 pKa = 9.27LLCDD715 pKa = 3.5ATDD718 pKa = 3.46TTDD721 pKa = 3.34CDD723 pKa = 3.61RR724 pKa = 11.84AKK726 pKa = 10.14VTISVYY732 pKa = 10.64DD733 pKa = 3.51GNGFDD738 pKa = 5.84FGDD741 pKa = 4.35APPSYY746 pKa = 9.23LTASHH751 pKa = 6.73GVPDD755 pKa = 3.98TPVIYY760 pKa = 10.48LGVVPPDD767 pKa = 3.61AEE769 pKa = 4.77LVAQSDD775 pKa = 3.93ATATADD781 pKa = 4.17DD782 pKa = 4.4VFNIDD787 pKa = 5.34DD788 pKa = 3.98EE789 pKa = 4.91DD790 pKa = 4.0AVSFPILTQGTISTLDD806 pKa = 3.17VGVTGNGTLQAWIDD820 pKa = 3.87FNGDD824 pKa = 3.3GLFEE828 pKa = 4.05QTLGEE833 pKa = 4.8RR834 pKa = 11.84IATDD838 pKa = 3.51LRR840 pKa = 11.84DD841 pKa = 4.26DD842 pKa = 4.36GSAFDD847 pKa = 4.63NVAGDD852 pKa = 3.85GVIQIDD858 pKa = 3.87VAVPTDD864 pKa = 3.4ATTDD868 pKa = 3.2TTFARR873 pKa = 11.84FRR875 pKa = 11.84YY876 pKa = 7.75STSPGLSPTDD886 pKa = 3.77FAPDD890 pKa = 3.92GEE892 pKa = 4.53VEE894 pKa = 4.61DD895 pKa = 4.39YY896 pKa = 11.45SLVIAAADD904 pKa = 3.6LVDD907 pKa = 5.34RR908 pKa = 11.84GDD910 pKa = 3.89APASYY915 pKa = 10.6GDD917 pKa = 3.68PRR919 pKa = 11.84HH920 pKa = 6.61IIVDD924 pKa = 4.47DD925 pKa = 3.73IFLGAGIPDD934 pKa = 4.54SEE936 pKa = 4.42TDD938 pKa = 3.5PQNSVDD944 pKa = 4.55ADD946 pKa = 3.73ADD948 pKa = 3.99DD949 pKa = 4.19FAGADD954 pKa = 4.15DD955 pKa = 4.43EE956 pKa = 5.92DD957 pKa = 4.53SVAVFPVFEE966 pKa = 4.35VGTTVSVTVQTHH978 pKa = 4.6EE979 pKa = 4.31TLSLQYY985 pKa = 11.49ALGIPVTSGVTNLQAWIDD1003 pKa = 3.7FDD1005 pKa = 4.34QNGTFEE1011 pKa = 4.48PSEE1014 pKa = 4.02QVATNYY1020 pKa = 10.17RR1021 pKa = 11.84DD1022 pKa = 3.49GGIGDD1027 pKa = 3.69TDD1029 pKa = 4.73GIFNNQISFDD1039 pKa = 3.68VTVPNTISSGYY1050 pKa = 8.49TYY1052 pKa = 11.42ARR1054 pKa = 11.84LRR1056 pKa = 11.84WSTSSALVSDD1066 pKa = 4.82PFDD1069 pKa = 3.53GLNFDD1074 pKa = 5.32GEE1076 pKa = 4.54VEE1078 pKa = 4.22DD1079 pKa = 4.61YY1080 pKa = 11.11EE1081 pKa = 4.75VVLSAGAVPFEE1092 pKa = 4.74CDD1094 pKa = 2.74GTLYY1098 pKa = 10.33RR1099 pKa = 11.84VARR1102 pKa = 11.84VDD1104 pKa = 3.36SQLQRR1109 pKa = 11.84LVFSEE1114 pKa = 4.78DD1115 pKa = 2.93GSGGYY1120 pKa = 8.7TIDD1123 pKa = 3.62ATNVGAPAGVAYY1135 pKa = 10.79NGGWGYY1141 pKa = 10.77NAVDD1145 pKa = 3.37GLFYY1149 pKa = 10.69GVRR1152 pKa = 11.84EE1153 pKa = 4.1FEE1155 pKa = 4.22RR1156 pKa = 11.84DD1157 pKa = 3.38LVQLDD1162 pKa = 3.61STGKK1166 pKa = 8.01FTVVSTLPPTAATGYY1181 pKa = 10.28NSGDD1185 pKa = 3.33ILSNGIMIYY1194 pKa = 10.11RR1195 pKa = 11.84VQGTNDD1201 pKa = 3.36FQLLDD1206 pKa = 3.57ITDD1209 pKa = 3.77PTNPIDD1215 pKa = 4.21AGLVTLTSAVDD1226 pKa = 3.57PFDD1229 pKa = 4.03IAFNPNDD1236 pKa = 3.21GMIYY1240 pKa = 10.21GVNHH1244 pKa = 4.57VTDD1247 pKa = 3.56RR1248 pKa = 11.84LFYY1251 pKa = 10.55FDD1253 pKa = 5.09PADD1256 pKa = 3.98GAPGTRR1262 pKa = 11.84TPVEE1266 pKa = 4.27FGPAIWTEE1274 pKa = 3.9AYY1276 pKa = 9.86GAIWFDD1282 pKa = 3.72FYY1284 pKa = 11.75GRR1286 pKa = 11.84MYY1288 pKa = 11.16VNQNTSNEE1296 pKa = 4.14MYY1298 pKa = 9.96EE1299 pKa = 4.04VDD1301 pKa = 5.34VGIQGNGTGNRR1312 pKa = 11.84EE1313 pKa = 4.64LISVITISEE1322 pKa = 4.07EE1323 pKa = 3.7FRR1325 pKa = 11.84NDD1327 pKa = 3.26GAGCPSRR1334 pKa = 11.84LGPLPPEE1341 pKa = 4.25GALAGTVYY1349 pKa = 10.87EE1350 pKa = 4.56DD1351 pKa = 3.9TDD1353 pKa = 3.91GSGAFDD1359 pKa = 3.57NGEE1362 pKa = 3.98TGIPNISVDD1371 pKa = 3.23IYY1373 pKa = 11.2NDD1375 pKa = 2.99NGTPGTIVDD1384 pKa = 4.32DD1385 pKa = 4.08VLVTSVVTAADD1396 pKa = 3.52GSYY1399 pKa = 10.62LVEE1402 pKa = 4.65NLSALNTYY1410 pKa = 10.23RR1411 pKa = 11.84IEE1413 pKa = 4.6VEE1415 pKa = 4.61DD1416 pKa = 4.96ADD1418 pKa = 4.41ADD1420 pKa = 3.99MPAGYY1425 pKa = 10.18QSVTPNPLTAIRR1437 pKa = 11.84VTAGSTRR1444 pKa = 11.84GGLDD1448 pKa = 3.13FGFAPGEE1455 pKa = 4.01ADD1457 pKa = 3.97LSITKK1462 pKa = 8.87TVEE1465 pKa = 3.87RR1466 pKa = 11.84TSDD1469 pKa = 3.29GATVTSAAEE1478 pKa = 4.1GTEE1481 pKa = 4.07LEE1483 pKa = 4.41FVISVTNSGPASVASVLVNDD1503 pKa = 4.68ILPSGFAYY1511 pKa = 10.76VSDD1514 pKa = 4.36DD1515 pKa = 3.92AASQGNTYY1523 pKa = 10.3NASTGRR1529 pKa = 11.84WDD1531 pKa = 3.56VGGLAIGEE1539 pKa = 4.47TKK1541 pKa = 9.38TLRR1544 pKa = 11.84ITVTMLGSGNHH1555 pKa = 4.91TNIAEE1560 pKa = 4.48IIFSSVDD1567 pKa = 3.83DD1568 pKa = 4.76PDD1570 pKa = 4.33SDD1572 pKa = 4.14PSVGTATDD1580 pKa = 4.13DD1581 pKa = 4.14LSDD1584 pKa = 5.47GIPDD1588 pKa = 4.14DD1589 pKa = 5.58DD1590 pKa = 4.07EE1591 pKa = 7.09ASVTVALDD1599 pKa = 3.16NGEE1602 pKa = 4.22RR1603 pKa = 11.84LLSGRR1608 pKa = 11.84VFIDD1612 pKa = 2.9NGVNSGTAHH1621 pKa = 6.74DD1622 pKa = 5.25AIVNGSEE1629 pKa = 4.19TGTQSATLTILDD1641 pKa = 3.89GSGSVIATPTIAADD1655 pKa = 4.13GSWAYY1660 pKa = 10.58ALDD1663 pKa = 3.98GSYY1666 pKa = 10.49SGPLSVSVAPLPGFITVSEE1685 pKa = 4.23NDD1687 pKa = 3.22SGLPALANSDD1697 pKa = 3.45PHH1699 pKa = 7.71DD1700 pKa = 4.32GNFTFTPDD1708 pKa = 2.93ASSNYY1713 pKa = 9.45SGLNIGIVRR1722 pKa = 11.84APTLTRR1728 pKa = 11.84DD1729 pKa = 3.32QEE1731 pKa = 4.22ASVEE1735 pKa = 4.25SGQVATLQHH1744 pKa = 7.28EE1745 pKa = 4.69YY1746 pKa = 9.84TAFTSGTVTFSYY1758 pKa = 10.82ANPNMAPPNSFSATLYY1774 pKa = 10.72RR1775 pKa = 11.84DD1776 pKa = 3.75TDD1778 pKa = 3.92CDD1780 pKa = 3.91TTPDD1784 pKa = 3.66TPINAPITVSAGDD1797 pKa = 3.8RR1798 pKa = 11.84LCLVSRR1804 pKa = 11.84VTANSGIGPGSSYY1817 pKa = 10.78TYY1819 pKa = 11.15DD1820 pKa = 3.52LVASTALSATTVVSTSQNTDD1840 pKa = 3.46RR1841 pKa = 11.84IVAGGGAGRR1850 pKa = 11.84LEE1852 pKa = 3.87LRR1854 pKa = 11.84KK1855 pKa = 7.74TVRR1858 pKa = 11.84NITQNTPEE1866 pKa = 4.26GASNGGAIGDD1876 pKa = 3.65VLRR1879 pKa = 11.84YY1880 pKa = 9.56QIYY1883 pKa = 10.51LRR1885 pKa = 11.84NPSSAPATNVIIYY1898 pKa = 9.45DD1899 pKa = 3.59KK1900 pKa = 9.93TPSYY1904 pKa = 9.52TALASPIASPTTLGNNLICVVSQPTNNVAGYY1935 pKa = 9.46VGPLRR1940 pKa = 11.84WDD1942 pKa = 3.32CTGTYY1947 pKa = 10.35EE1948 pKa = 4.92PGEE1951 pKa = 4.45DD1952 pKa = 3.26DD1953 pKa = 4.73SVFFDD1958 pKa = 4.36VVISPP1963 pKa = 4.35
Molecular weight: 204.57 kDa
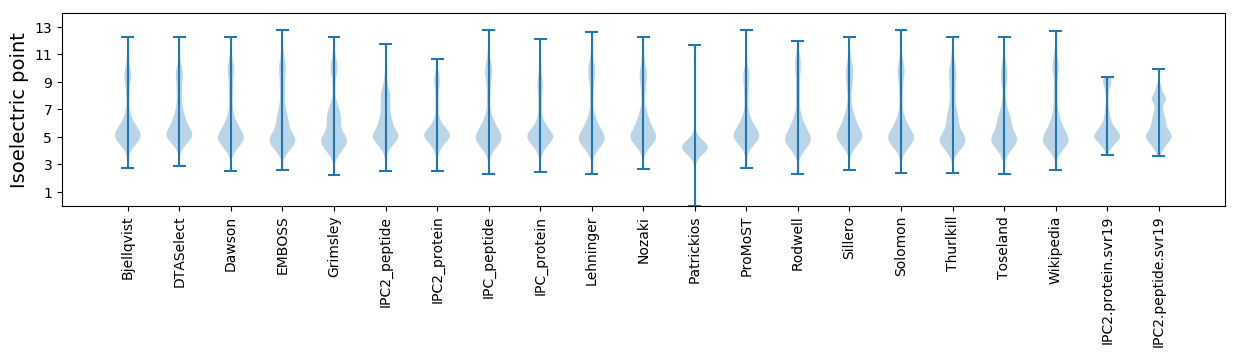
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F4RL47|A0A0F4RL47_9RHOB Uncharacterized protein OS=Loktanella sp. S4079 OX=579483 GN=TW80_05690 PE=4 SV=1
MM1 pKa = 6.48TQRR4 pKa = 11.84VDD6 pKa = 3.31VAANRR11 pKa = 11.84AARR14 pKa = 11.84KK15 pKa = 9.54YY16 pKa = 10.61DD17 pKa = 3.11ALTQVKK23 pKa = 9.31RR24 pKa = 11.84VLWALCWPLFRR35 pKa = 11.84LSPRR39 pKa = 11.84PMWGWRR45 pKa = 11.84VWILRR50 pKa = 11.84AFGAQVGRR58 pKa = 11.84GVHH61 pKa = 6.0VYY63 pKa = 9.09PSARR67 pKa = 11.84ITMPWHH73 pKa = 6.31LRR75 pKa = 11.84LADD78 pKa = 3.7EE79 pKa = 4.38VAIGDD84 pKa = 3.93RR85 pKa = 11.84AILYY89 pKa = 9.96ALDD92 pKa = 4.22VITIGARR99 pKa = 11.84ATVSQYY105 pKa = 11.68AHH107 pKa = 7.41LCTGSHH113 pKa = 7.3DD114 pKa = 4.21LGDD117 pKa = 3.68PARR120 pKa = 11.84ALKK123 pKa = 8.46TAPIMIGADD132 pKa = 2.78AWVCADD138 pKa = 3.66AFVGPGVQIGKK149 pKa = 9.28SAVLGARR156 pKa = 11.84AVAFKK161 pKa = 10.58ALPDD165 pKa = 3.46RR166 pKa = 11.84CVGIGNPMQIRR177 pKa = 11.84SRR179 pKa = 11.84HH180 pKa = 5.08DD181 pKa = 2.95
MM1 pKa = 6.48TQRR4 pKa = 11.84VDD6 pKa = 3.31VAANRR11 pKa = 11.84AARR14 pKa = 11.84KK15 pKa = 9.54YY16 pKa = 10.61DD17 pKa = 3.11ALTQVKK23 pKa = 9.31RR24 pKa = 11.84VLWALCWPLFRR35 pKa = 11.84LSPRR39 pKa = 11.84PMWGWRR45 pKa = 11.84VWILRR50 pKa = 11.84AFGAQVGRR58 pKa = 11.84GVHH61 pKa = 6.0VYY63 pKa = 9.09PSARR67 pKa = 11.84ITMPWHH73 pKa = 6.31LRR75 pKa = 11.84LADD78 pKa = 3.7EE79 pKa = 4.38VAIGDD84 pKa = 3.93RR85 pKa = 11.84AILYY89 pKa = 9.96ALDD92 pKa = 4.22VITIGARR99 pKa = 11.84ATVSQYY105 pKa = 11.68AHH107 pKa = 7.41LCTGSHH113 pKa = 7.3DD114 pKa = 4.21LGDD117 pKa = 3.68PARR120 pKa = 11.84ALKK123 pKa = 8.46TAPIMIGADD132 pKa = 2.78AWVCADD138 pKa = 3.66AFVGPGVQIGKK149 pKa = 9.28SAVLGARR156 pKa = 11.84AVAFKK161 pKa = 10.58ALPDD165 pKa = 3.46RR166 pKa = 11.84CVGIGNPMQIRR177 pKa = 11.84SRR179 pKa = 11.84HH180 pKa = 5.08DD181 pKa = 2.95
Molecular weight: 19.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1023294 |
33 |
1963 |
310.5 |
33.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.625 ± 0.051 | 0.911 ± 0.013 |
6.269 ± 0.037 | 5.726 ± 0.042 |
3.859 ± 0.031 | 8.338 ± 0.047 |
2.074 ± 0.022 | 5.788 ± 0.031 |
3.447 ± 0.038 | 9.6 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.827 ± 0.02 | 3.067 ± 0.021 |
4.765 ± 0.028 | 3.558 ± 0.03 |
5.943 ± 0.041 | 5.376 ± 0.027 |
5.77 ± 0.032 | 7.33 ± 0.039 |
1.365 ± 0.018 | 2.363 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
