
Ceratitis capitata sigmavirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Sigmavirus; Capitata sigmavirus
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
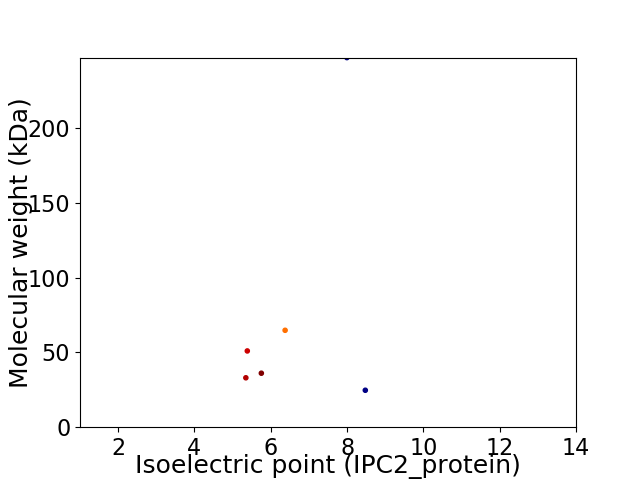
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140D8P3|A0A140D8P3_9RHAB Polymerase-associated protein OS=Ceratitis capitata sigmavirus OX=1802949 GN=P PE=4 SV=1
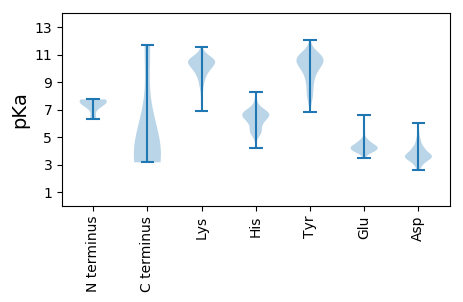
MM1 pKa = 6.33TTNGKK6 pKa = 10.18KK7 pKa = 9.62IVNINSKK14 pKa = 10.01KK15 pKa = 10.36SYY17 pKa = 10.29VVPCVYY23 pKa = 10.72AEE25 pKa = 4.31RR26 pKa = 11.84SPEE29 pKa = 4.25FPIDD33 pKa = 3.25WFDD36 pKa = 3.44TTRR39 pKa = 11.84DD40 pKa = 3.63KK41 pKa = 11.14PILNIQIFKK50 pKa = 10.83EE51 pKa = 3.95CDD53 pKa = 2.99LDD55 pKa = 4.24KK56 pKa = 11.26ARR58 pKa = 11.84QYY60 pKa = 12.09ADD62 pKa = 3.7AFLKK66 pKa = 9.57GTIHH70 pKa = 5.64GTIPVTTYY78 pKa = 10.9LYY80 pKa = 11.22YY81 pKa = 11.2LFLTAKK87 pKa = 7.73EE88 pKa = 4.16TLTRR92 pKa = 11.84DD93 pKa = 2.87WTSYY97 pKa = 10.35RR98 pKa = 11.84MNLKK102 pKa = 10.31ASEE105 pKa = 4.18QVTPLSLLTVNKK117 pKa = 10.3EE118 pKa = 4.3EE119 pKa = 4.31VDD121 pKa = 3.54QTPLTNPTTLDD132 pKa = 3.51NNSDD136 pKa = 3.5KK137 pKa = 11.18SILLALVGIYY147 pKa = 9.92RR148 pKa = 11.84LHH150 pKa = 5.45TTHH153 pKa = 7.45PALVDD158 pKa = 3.32IVTDD162 pKa = 4.94RR163 pKa = 11.84INLLIQQATPSDD175 pKa = 3.53KK176 pKa = 10.74VQYY179 pKa = 10.84SVDD182 pKa = 3.66LAKK185 pKa = 10.34TNSGYY190 pKa = 11.09LSGNDD195 pKa = 3.21SVEE198 pKa = 4.13ILLSALDD205 pKa = 3.64MFADD209 pKa = 4.18KK210 pKa = 10.85FPANKK215 pKa = 9.85YY216 pKa = 7.67SQARR220 pKa = 11.84IGTIILRR227 pKa = 11.84YY228 pKa = 9.21AGCSALLDD236 pKa = 3.8LTYY239 pKa = 7.27MTKK242 pKa = 9.92MIACDD247 pKa = 3.46GVLDD251 pKa = 3.74VLQWVFLPRR260 pKa = 11.84VGQEE264 pKa = 3.64LDD266 pKa = 3.24AMLSKK271 pKa = 10.67EE272 pKa = 3.92DD273 pKa = 4.04SEE275 pKa = 4.36ITKK278 pKa = 10.07EE279 pKa = 4.1DD280 pKa = 3.75SYY282 pKa = 11.77FPYY285 pKa = 11.05LLGLRR290 pKa = 11.84LSSKK294 pKa = 10.5SPYY297 pKa = 9.26AASSAPQLHH306 pKa = 6.5HH307 pKa = 6.87LVHH310 pKa = 6.7AVGSLMGLSRR320 pKa = 11.84SINALLIDD328 pKa = 4.46PGTPNMVANNAALIFLANKK347 pKa = 9.5RR348 pKa = 11.84LSGLKK353 pKa = 9.29VVYY356 pKa = 10.05MNEE359 pKa = 3.8DD360 pKa = 3.54DD361 pKa = 5.14AKK363 pKa = 11.26VNQTQQEE370 pKa = 4.48KK371 pKa = 10.68ASQTRR376 pKa = 11.84QQNISEE382 pKa = 4.09EE383 pKa = 4.4DD384 pKa = 3.46ALSSRR389 pKa = 11.84DD390 pKa = 4.4LDD392 pKa = 4.19DD393 pKa = 4.84EE394 pKa = 4.44QPKK397 pKa = 9.0TPRR400 pKa = 11.84DD401 pKa = 3.3WFNWYY406 pKa = 9.61CDD408 pKa = 4.05KK409 pKa = 11.04DD410 pKa = 3.45WKK412 pKa = 9.21FTKK415 pKa = 10.14KK416 pKa = 10.15EE417 pKa = 3.76YY418 pKa = 11.11LEE420 pKa = 4.2IRR422 pKa = 11.84DD423 pKa = 3.54AVMSIKK429 pKa = 10.4NPRR432 pKa = 11.84SGTVGAWTVEE442 pKa = 4.27TFLSLINADD451 pKa = 3.46IYY453 pKa = 11.7
MM1 pKa = 6.33TTNGKK6 pKa = 10.18KK7 pKa = 9.62IVNINSKK14 pKa = 10.01KK15 pKa = 10.36SYY17 pKa = 10.29VVPCVYY23 pKa = 10.72AEE25 pKa = 4.31RR26 pKa = 11.84SPEE29 pKa = 4.25FPIDD33 pKa = 3.25WFDD36 pKa = 3.44TTRR39 pKa = 11.84DD40 pKa = 3.63KK41 pKa = 11.14PILNIQIFKK50 pKa = 10.83EE51 pKa = 3.95CDD53 pKa = 2.99LDD55 pKa = 4.24KK56 pKa = 11.26ARR58 pKa = 11.84QYY60 pKa = 12.09ADD62 pKa = 3.7AFLKK66 pKa = 9.57GTIHH70 pKa = 5.64GTIPVTTYY78 pKa = 10.9LYY80 pKa = 11.22YY81 pKa = 11.2LFLTAKK87 pKa = 7.73EE88 pKa = 4.16TLTRR92 pKa = 11.84DD93 pKa = 2.87WTSYY97 pKa = 10.35RR98 pKa = 11.84MNLKK102 pKa = 10.31ASEE105 pKa = 4.18QVTPLSLLTVNKK117 pKa = 10.3EE118 pKa = 4.3EE119 pKa = 4.31VDD121 pKa = 3.54QTPLTNPTTLDD132 pKa = 3.51NNSDD136 pKa = 3.5KK137 pKa = 11.18SILLALVGIYY147 pKa = 9.92RR148 pKa = 11.84LHH150 pKa = 5.45TTHH153 pKa = 7.45PALVDD158 pKa = 3.32IVTDD162 pKa = 4.94RR163 pKa = 11.84INLLIQQATPSDD175 pKa = 3.53KK176 pKa = 10.74VQYY179 pKa = 10.84SVDD182 pKa = 3.66LAKK185 pKa = 10.34TNSGYY190 pKa = 11.09LSGNDD195 pKa = 3.21SVEE198 pKa = 4.13ILLSALDD205 pKa = 3.64MFADD209 pKa = 4.18KK210 pKa = 10.85FPANKK215 pKa = 9.85YY216 pKa = 7.67SQARR220 pKa = 11.84IGTIILRR227 pKa = 11.84YY228 pKa = 9.21AGCSALLDD236 pKa = 3.8LTYY239 pKa = 7.27MTKK242 pKa = 9.92MIACDD247 pKa = 3.46GVLDD251 pKa = 3.74VLQWVFLPRR260 pKa = 11.84VGQEE264 pKa = 3.64LDD266 pKa = 3.24AMLSKK271 pKa = 10.67EE272 pKa = 3.92DD273 pKa = 4.04SEE275 pKa = 4.36ITKK278 pKa = 10.07EE279 pKa = 4.1DD280 pKa = 3.75SYY282 pKa = 11.77FPYY285 pKa = 11.05LLGLRR290 pKa = 11.84LSSKK294 pKa = 10.5SPYY297 pKa = 9.26AASSAPQLHH306 pKa = 6.5HH307 pKa = 6.87LVHH310 pKa = 6.7AVGSLMGLSRR320 pKa = 11.84SINALLIDD328 pKa = 4.46PGTPNMVANNAALIFLANKK347 pKa = 9.5RR348 pKa = 11.84LSGLKK353 pKa = 9.29VVYY356 pKa = 10.05MNEE359 pKa = 3.8DD360 pKa = 3.54DD361 pKa = 5.14AKK363 pKa = 11.26VNQTQQEE370 pKa = 4.48KK371 pKa = 10.68ASQTRR376 pKa = 11.84QQNISEE382 pKa = 4.09EE383 pKa = 4.4DD384 pKa = 3.46ALSSRR389 pKa = 11.84DD390 pKa = 4.4LDD392 pKa = 4.19DD393 pKa = 4.84EE394 pKa = 4.44QPKK397 pKa = 9.0TPRR400 pKa = 11.84DD401 pKa = 3.3WFNWYY406 pKa = 9.61CDD408 pKa = 4.05KK409 pKa = 11.04DD410 pKa = 3.45WKK412 pKa = 9.21FTKK415 pKa = 10.14KK416 pKa = 10.15EE417 pKa = 3.76YY418 pKa = 11.11LEE420 pKa = 4.2IRR422 pKa = 11.84DD423 pKa = 3.54AVMSIKK429 pKa = 10.4NPRR432 pKa = 11.84SGTVGAWTVEE442 pKa = 4.27TFLSLINADD451 pKa = 3.46IYY453 pKa = 11.7
Molecular weight: 51.03 kDa
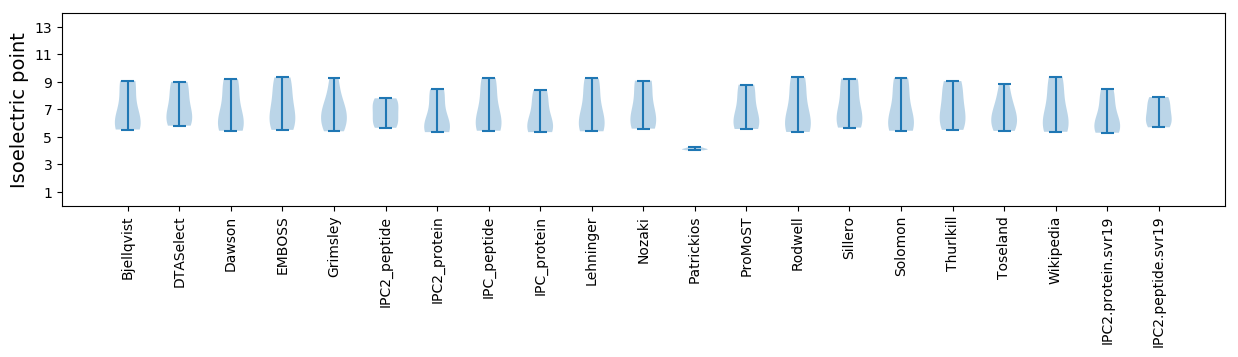
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140D8P6|A0A140D8P6_9RHAB Glycoprotein OS=Ceratitis capitata sigmavirus OX=1802949 GN=G PE=4 SV=1
MM1 pKa = 7.39NGKK4 pKa = 8.81KK5 pKa = 9.91QLVRR9 pKa = 11.84FKK11 pKa = 10.16EE12 pKa = 4.08QTPTTSKK19 pKa = 10.88NLSKK23 pKa = 10.45PKK25 pKa = 10.47EE26 pKa = 4.26EE27 pKa = 3.89ISSPFKK33 pKa = 10.52PIYY36 pKa = 7.59TTTLPSAPTLTPKK49 pKa = 9.91VIKK52 pKa = 9.96WDD54 pKa = 3.6VTSSLRR60 pKa = 11.84LTTNRR65 pKa = 11.84RR66 pKa = 11.84PKK68 pKa = 8.78EE69 pKa = 3.96WKK71 pKa = 10.1SIQCILAVLIDD82 pKa = 4.62EE83 pKa = 4.6YY84 pKa = 11.17AGPEE88 pKa = 3.53MWRR91 pKa = 11.84SVFEE95 pKa = 3.88LFYY98 pKa = 10.99WMLSRR103 pKa = 11.84NIAEE107 pKa = 4.9LPSSSDD113 pKa = 3.13RR114 pKa = 11.84YY115 pKa = 11.02VYY117 pKa = 10.7GADD120 pKa = 3.61FNEE123 pKa = 4.36LTLVHH128 pKa = 6.6HH129 pKa = 6.95NIYY132 pKa = 10.84YY133 pKa = 10.78LIDD136 pKa = 3.35QGQDD140 pKa = 3.13YY141 pKa = 10.09HH142 pKa = 6.44FHH144 pKa = 5.34YY145 pKa = 9.49TGKK148 pKa = 9.79IRR150 pKa = 11.84DD151 pKa = 3.76MKK153 pKa = 10.2YY154 pKa = 10.58SIVFSCSLKK163 pKa = 8.58PTKK166 pKa = 9.98RR167 pKa = 11.84DD168 pKa = 3.41CYY170 pKa = 10.83SIARR174 pKa = 11.84LQEE177 pKa = 4.01YY178 pKa = 10.45KK179 pKa = 10.84SIIYY183 pKa = 10.24EE184 pKa = 4.71DD185 pKa = 4.0IPDD188 pKa = 3.93LTTVLTLGQVKK199 pKa = 10.42YY200 pKa = 10.78MMLNQTIVLHH210 pKa = 5.35MM211 pKa = 4.68
MM1 pKa = 7.39NGKK4 pKa = 8.81KK5 pKa = 9.91QLVRR9 pKa = 11.84FKK11 pKa = 10.16EE12 pKa = 4.08QTPTTSKK19 pKa = 10.88NLSKK23 pKa = 10.45PKK25 pKa = 10.47EE26 pKa = 4.26EE27 pKa = 3.89ISSPFKK33 pKa = 10.52PIYY36 pKa = 7.59TTTLPSAPTLTPKK49 pKa = 9.91VIKK52 pKa = 9.96WDD54 pKa = 3.6VTSSLRR60 pKa = 11.84LTTNRR65 pKa = 11.84RR66 pKa = 11.84PKK68 pKa = 8.78EE69 pKa = 3.96WKK71 pKa = 10.1SIQCILAVLIDD82 pKa = 4.62EE83 pKa = 4.6YY84 pKa = 11.17AGPEE88 pKa = 3.53MWRR91 pKa = 11.84SVFEE95 pKa = 3.88LFYY98 pKa = 10.99WMLSRR103 pKa = 11.84NIAEE107 pKa = 4.9LPSSSDD113 pKa = 3.13RR114 pKa = 11.84YY115 pKa = 11.02VYY117 pKa = 10.7GADD120 pKa = 3.61FNEE123 pKa = 4.36LTLVHH128 pKa = 6.6HH129 pKa = 6.95NIYY132 pKa = 10.84YY133 pKa = 10.78LIDD136 pKa = 3.35QGQDD140 pKa = 3.13YY141 pKa = 10.09HH142 pKa = 6.44FHH144 pKa = 5.34YY145 pKa = 9.49TGKK148 pKa = 9.79IRR150 pKa = 11.84DD151 pKa = 3.76MKK153 pKa = 10.2YY154 pKa = 10.58SIVFSCSLKK163 pKa = 8.58PTKK166 pKa = 9.98RR167 pKa = 11.84DD168 pKa = 3.41CYY170 pKa = 10.83SIARR174 pKa = 11.84LQEE177 pKa = 4.01YY178 pKa = 10.45KK179 pKa = 10.84SIIYY183 pKa = 10.24EE184 pKa = 4.71DD185 pKa = 4.0IPDD188 pKa = 3.93LTTVLTLGQVKK199 pKa = 10.42YY200 pKa = 10.78MMLNQTIVLHH210 pKa = 5.35MM211 pKa = 4.68
Molecular weight: 24.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3981 |
211 |
2152 |
663.5 |
76.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.391 ± 0.596 | 1.658 ± 0.151 |
5.526 ± 0.587 | 4.999 ± 0.223 |
4.295 ± 0.421 | 4.446 ± 0.539 |
2.914 ± 0.28 | 8.415 ± 0.481 |
6.33 ± 0.23 | 10.952 ± 0.249 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.035 ± 0.226 | 6.556 ± 0.35 |
4.496 ± 0.308 | 3.542 ± 0.273 |
4.521 ± 0.192 | 8.314 ± 0.274 |
6.883 ± 0.279 | 4.974 ± 0.402 |
1.557 ± 0.267 | 4.195 ± 0.248 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
