
Oceanimonas sp. (strain GK1 / IBRC-M 10197)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Aeromonadales; Aeromonadaceae; Oceanimonas; unclassified Oceanimonas
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
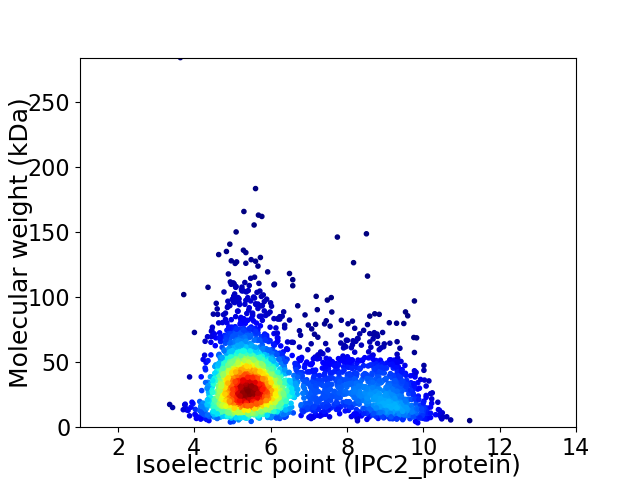
Virtual 2D-PAGE plot for 3190 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H2FWV9|H2FWV9_OCESG Lipopolysaccharide assembly protein B OS=Oceanimonas sp. (strain GK1 / IBRC-M 10197) OX=511062 GN=lapB PE=3 SV=1
MM1 pKa = 7.68KK2 pKa = 9.38KK3 pKa = 9.08TILALTIPALFATSASAVTVYY24 pKa = 10.62SDD26 pKa = 3.61EE27 pKa = 4.27GAQVDD32 pKa = 3.41IYY34 pKa = 11.54GRR36 pKa = 11.84VQFDD40 pKa = 3.28GGEE43 pKa = 4.17LDD45 pKa = 4.27HH46 pKa = 7.83QNDD49 pKa = 4.13SNGKK53 pKa = 6.77EE54 pKa = 4.12VKK56 pKa = 10.19SEE58 pKa = 4.1SFGTDD63 pKa = 2.56GSARR67 pKa = 11.84LGVNMSYY74 pKa = 11.5ALNNDD79 pKa = 2.44VDD81 pKa = 5.52LIGKK85 pKa = 8.14LEE87 pKa = 3.96WQVAAEE93 pKa = 4.49ASDD96 pKa = 3.79DD97 pKa = 4.05SKK99 pKa = 11.67FDD101 pKa = 4.46ARR103 pKa = 11.84YY104 pKa = 8.28AWAGFRR110 pKa = 11.84FMDD113 pKa = 3.7TTEE116 pKa = 4.0LTFGRR121 pKa = 11.84SVDD124 pKa = 3.76PLAQAIYY131 pKa = 8.87LTDD134 pKa = 3.23VFNIFGAGITYY145 pKa = 10.21GSEE148 pKa = 3.78FSISDD153 pKa = 3.6KK154 pKa = 11.43ADD156 pKa = 3.48DD157 pKa = 4.77QIMATYY163 pKa = 9.53AANGVDD169 pKa = 4.62LRR171 pKa = 11.84AAYY174 pKa = 10.09AFADD178 pKa = 4.12DD179 pKa = 5.3DD180 pKa = 3.88RR181 pKa = 11.84TDD183 pKa = 3.91FANQANGDD191 pKa = 3.68VAEE194 pKa = 4.37NQWAVSAGYY203 pKa = 7.48TFPFGLGLVAAYY215 pKa = 7.42EE216 pKa = 4.06QQNGYY221 pKa = 9.75EE222 pKa = 4.33KK223 pKa = 10.37TGLTSQNIDD232 pKa = 2.94QDD234 pKa = 3.02VWYY237 pKa = 10.45AGVHH241 pKa = 4.1YY242 pKa = 9.04TLDD245 pKa = 3.35GFYY248 pKa = 10.51FAALYY253 pKa = 10.55SDD255 pKa = 4.03RR256 pKa = 11.84QRR258 pKa = 11.84DD259 pKa = 3.45TGTAAGDD266 pKa = 4.03DD267 pKa = 3.68EE268 pKa = 4.45GTAYY272 pKa = 9.95EE273 pKa = 4.8LHH275 pKa = 6.15AQYY278 pKa = 11.21NVDD281 pKa = 3.51AWTLMAQYY289 pKa = 10.69SKK291 pKa = 11.59EE292 pKa = 4.09EE293 pKa = 3.8FDD295 pKa = 3.62AAGATEE301 pKa = 4.96EE302 pKa = 4.58YY303 pKa = 11.0DD304 pKa = 3.95SIDD307 pKa = 4.97DD308 pKa = 3.53ITLGVQYY315 pKa = 11.16DD316 pKa = 4.09LTSKK320 pKa = 9.03TKK322 pKa = 10.55LYY324 pKa = 10.83AEE326 pKa = 4.29YY327 pKa = 10.29VISDD331 pKa = 4.25SEE333 pKa = 4.35VDD335 pKa = 3.16TSGAKK340 pKa = 9.8KK341 pKa = 10.35DD342 pKa = 3.65DD343 pKa = 4.02LYY345 pKa = 11.73GVGIQYY351 pKa = 10.53NFF353 pKa = 3.35
MM1 pKa = 7.68KK2 pKa = 9.38KK3 pKa = 9.08TILALTIPALFATSASAVTVYY24 pKa = 10.62SDD26 pKa = 3.61EE27 pKa = 4.27GAQVDD32 pKa = 3.41IYY34 pKa = 11.54GRR36 pKa = 11.84VQFDD40 pKa = 3.28GGEE43 pKa = 4.17LDD45 pKa = 4.27HH46 pKa = 7.83QNDD49 pKa = 4.13SNGKK53 pKa = 6.77EE54 pKa = 4.12VKK56 pKa = 10.19SEE58 pKa = 4.1SFGTDD63 pKa = 2.56GSARR67 pKa = 11.84LGVNMSYY74 pKa = 11.5ALNNDD79 pKa = 2.44VDD81 pKa = 5.52LIGKK85 pKa = 8.14LEE87 pKa = 3.96WQVAAEE93 pKa = 4.49ASDD96 pKa = 3.79DD97 pKa = 4.05SKK99 pKa = 11.67FDD101 pKa = 4.46ARR103 pKa = 11.84YY104 pKa = 8.28AWAGFRR110 pKa = 11.84FMDD113 pKa = 3.7TTEE116 pKa = 4.0LTFGRR121 pKa = 11.84SVDD124 pKa = 3.76PLAQAIYY131 pKa = 8.87LTDD134 pKa = 3.23VFNIFGAGITYY145 pKa = 10.21GSEE148 pKa = 3.78FSISDD153 pKa = 3.6KK154 pKa = 11.43ADD156 pKa = 3.48DD157 pKa = 4.77QIMATYY163 pKa = 9.53AANGVDD169 pKa = 4.62LRR171 pKa = 11.84AAYY174 pKa = 10.09AFADD178 pKa = 4.12DD179 pKa = 5.3DD180 pKa = 3.88RR181 pKa = 11.84TDD183 pKa = 3.91FANQANGDD191 pKa = 3.68VAEE194 pKa = 4.37NQWAVSAGYY203 pKa = 7.48TFPFGLGLVAAYY215 pKa = 7.42EE216 pKa = 4.06QQNGYY221 pKa = 9.75EE222 pKa = 4.33KK223 pKa = 10.37TGLTSQNIDD232 pKa = 2.94QDD234 pKa = 3.02VWYY237 pKa = 10.45AGVHH241 pKa = 4.1YY242 pKa = 9.04TLDD245 pKa = 3.35GFYY248 pKa = 10.51FAALYY253 pKa = 10.55SDD255 pKa = 4.03RR256 pKa = 11.84QRR258 pKa = 11.84DD259 pKa = 3.45TGTAAGDD266 pKa = 4.03DD267 pKa = 3.68EE268 pKa = 4.45GTAYY272 pKa = 9.95EE273 pKa = 4.8LHH275 pKa = 6.15AQYY278 pKa = 11.21NVDD281 pKa = 3.51AWTLMAQYY289 pKa = 10.69SKK291 pKa = 11.59EE292 pKa = 4.09EE293 pKa = 3.8FDD295 pKa = 3.62AAGATEE301 pKa = 4.96EE302 pKa = 4.58YY303 pKa = 11.0DD304 pKa = 3.95SIDD307 pKa = 4.97DD308 pKa = 3.53ITLGVQYY315 pKa = 11.16DD316 pKa = 4.09LTSKK320 pKa = 9.03TKK322 pKa = 10.55LYY324 pKa = 10.83AEE326 pKa = 4.29YY327 pKa = 10.29VISDD331 pKa = 4.25SEE333 pKa = 4.35VDD335 pKa = 3.16TSGAKK340 pKa = 9.8KK341 pKa = 10.35DD342 pKa = 3.65DD343 pKa = 4.02LYY345 pKa = 11.73GVGIQYY351 pKa = 10.53NFF353 pKa = 3.35
Molecular weight: 38.61 kDa
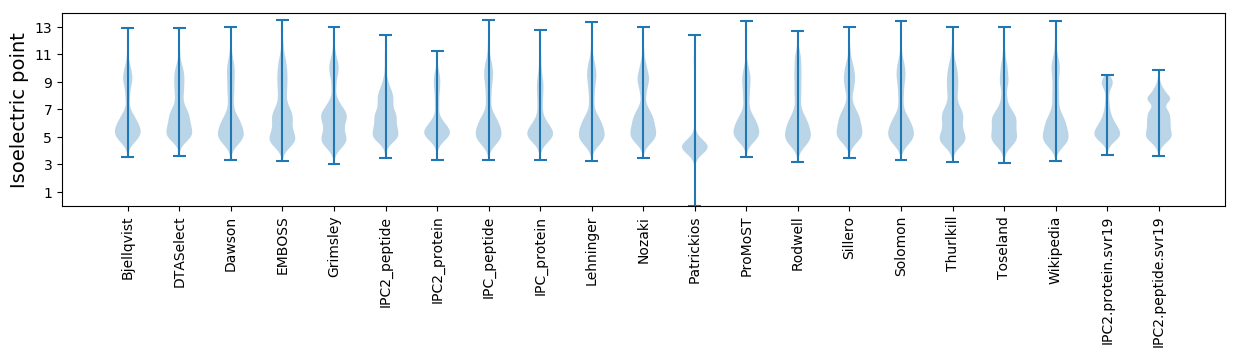
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H2FYZ9|H2FYZ9_OCESG RNA-binding protein Hfq OS=Oceanimonas sp. (strain GK1 / IBRC-M 10197) OX=511062 GN=hfq PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 9.13RR14 pKa = 11.84SHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 9.16VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.29GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 9.13RR14 pKa = 11.84SHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 9.16VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.29GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1032523 |
32 |
2846 |
323.7 |
35.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.771 ± 0.055 | 1.021 ± 0.014 |
5.226 ± 0.032 | 6.145 ± 0.039 |
3.587 ± 0.031 | 8.029 ± 0.042 |
2.477 ± 0.022 | 4.727 ± 0.036 |
3.293 ± 0.038 | 12.149 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.552 ± 0.022 | 3.041 ± 0.02 |
4.713 ± 0.028 | 4.668 ± 0.04 |
6.547 ± 0.048 | 5.263 ± 0.033 |
4.849 ± 0.034 | 7.009 ± 0.04 |
1.418 ± 0.018 | 2.515 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
