
Marinobacterium lutimaris
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Oceanospirillaceae; Marinobacterium
Average proteome isoelectric point is 5.89
Get precalculated fractions of proteins
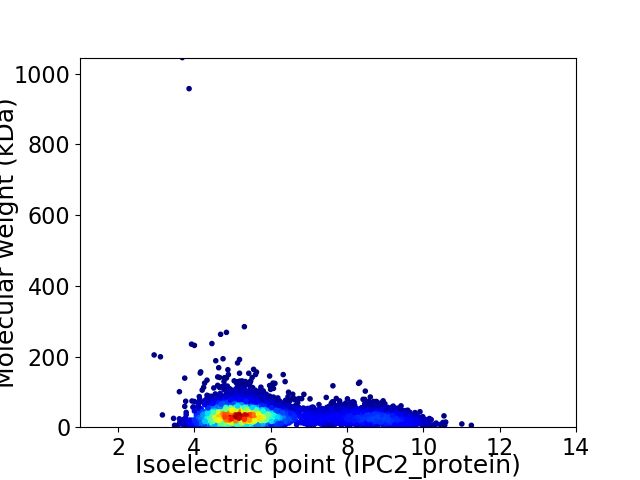
Virtual 2D-PAGE plot for 5061 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H5UWV7|A0A1H5UWV7_9GAMM Transglutaminase-like enzyme putative cysteine protease OS=Marinobacterium lutimaris OX=568106 GN=SAMN05444390_101544 PE=4 SV=1
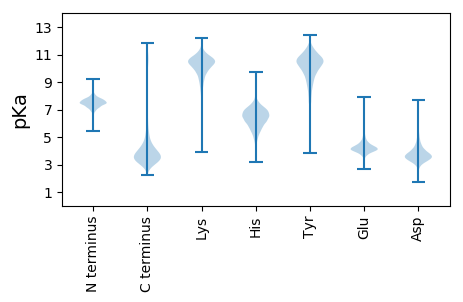
MM1 pKa = 7.4KK2 pKa = 10.32KK3 pKa = 10.35SLIALAVAGAMTAPVVAQADD23 pKa = 3.53ATLYY27 pKa = 11.08GNVEE31 pKa = 3.95IEE33 pKa = 4.3GVFANDD39 pKa = 3.2ADD41 pKa = 5.02AEE43 pKa = 4.31IQVDD47 pKa = 3.88DD48 pKa = 4.19ARR50 pKa = 11.84MGVKK54 pKa = 10.47GSDD57 pKa = 2.9EE58 pKa = 4.31TYY60 pKa = 10.27IDD62 pKa = 4.19GVSSFYY68 pKa = 10.76QIEE71 pKa = 4.44LEE73 pKa = 4.19YY74 pKa = 11.32NPDD77 pKa = 3.51SVLSDD82 pKa = 3.55GNSVTVRR89 pKa = 11.84KK90 pKa = 9.94ASAGLTGGFGTVIGGRR106 pKa = 11.84FSNPVEE112 pKa = 4.22STEE115 pKa = 5.02LNDD118 pKa = 4.9LYY120 pKa = 11.25SEE122 pKa = 4.26SLSTDD127 pKa = 3.94FFFRR131 pKa = 11.84DD132 pKa = 3.45PDD134 pKa = 4.25RR135 pKa = 11.84IGSALAYY142 pKa = 7.33ITPTFGGFSAYY153 pKa = 10.4AGIAADD159 pKa = 4.08GNADD163 pKa = 3.42RR164 pKa = 11.84HH165 pKa = 5.86ALNGADD171 pKa = 4.83NDD173 pKa = 4.25RR174 pKa = 11.84EE175 pKa = 4.15DD176 pKa = 4.23LDD178 pKa = 4.68GYY180 pKa = 11.18LVGADD185 pKa = 3.49YY186 pKa = 10.13TIGGFGAHH194 pKa = 6.53LGYY197 pKa = 10.47WSMDD201 pKa = 3.02KK202 pKa = 11.3DD203 pKa = 3.96GDD205 pKa = 4.19SEE207 pKa = 4.42AAGAAHH213 pKa = 6.81DD214 pKa = 4.11AEE216 pKa = 4.94YY217 pKa = 10.48IGLALSYY224 pKa = 10.38AVSNFTFTGSYY235 pKa = 11.04AEE237 pKa = 4.59ADD239 pKa = 3.79SVDD242 pKa = 3.32GGFIAGTVNTEE253 pKa = 3.59LWTLAADD260 pKa = 3.75YY261 pKa = 11.65AMEE264 pKa = 4.19NTNFGISYY272 pKa = 9.4MDD274 pKa = 3.4YY275 pKa = 10.99QEE277 pKa = 4.29SQKK280 pKa = 11.46SNDD283 pKa = 2.75IDD285 pKa = 4.0ADD287 pKa = 3.32EE288 pKa = 4.32WGVYY292 pKa = 9.34VSHH295 pKa = 7.13KK296 pKa = 10.25LSNKK300 pKa = 9.87ASVKK304 pKa = 10.14AQYY307 pKa = 10.12TSADD311 pKa = 3.25VDD313 pKa = 3.46NTDD316 pKa = 3.46IVGEE320 pKa = 4.13DD321 pKa = 3.75VFVVGYY327 pKa = 9.93NVSFF331 pKa = 4.04
MM1 pKa = 7.4KK2 pKa = 10.32KK3 pKa = 10.35SLIALAVAGAMTAPVVAQADD23 pKa = 3.53ATLYY27 pKa = 11.08GNVEE31 pKa = 3.95IEE33 pKa = 4.3GVFANDD39 pKa = 3.2ADD41 pKa = 5.02AEE43 pKa = 4.31IQVDD47 pKa = 3.88DD48 pKa = 4.19ARR50 pKa = 11.84MGVKK54 pKa = 10.47GSDD57 pKa = 2.9EE58 pKa = 4.31TYY60 pKa = 10.27IDD62 pKa = 4.19GVSSFYY68 pKa = 10.76QIEE71 pKa = 4.44LEE73 pKa = 4.19YY74 pKa = 11.32NPDD77 pKa = 3.51SVLSDD82 pKa = 3.55GNSVTVRR89 pKa = 11.84KK90 pKa = 9.94ASAGLTGGFGTVIGGRR106 pKa = 11.84FSNPVEE112 pKa = 4.22STEE115 pKa = 5.02LNDD118 pKa = 4.9LYY120 pKa = 11.25SEE122 pKa = 4.26SLSTDD127 pKa = 3.94FFFRR131 pKa = 11.84DD132 pKa = 3.45PDD134 pKa = 4.25RR135 pKa = 11.84IGSALAYY142 pKa = 7.33ITPTFGGFSAYY153 pKa = 10.4AGIAADD159 pKa = 4.08GNADD163 pKa = 3.42RR164 pKa = 11.84HH165 pKa = 5.86ALNGADD171 pKa = 4.83NDD173 pKa = 4.25RR174 pKa = 11.84EE175 pKa = 4.15DD176 pKa = 4.23LDD178 pKa = 4.68GYY180 pKa = 11.18LVGADD185 pKa = 3.49YY186 pKa = 10.13TIGGFGAHH194 pKa = 6.53LGYY197 pKa = 10.47WSMDD201 pKa = 3.02KK202 pKa = 11.3DD203 pKa = 3.96GDD205 pKa = 4.19SEE207 pKa = 4.42AAGAAHH213 pKa = 6.81DD214 pKa = 4.11AEE216 pKa = 4.94YY217 pKa = 10.48IGLALSYY224 pKa = 10.38AVSNFTFTGSYY235 pKa = 11.04AEE237 pKa = 4.59ADD239 pKa = 3.79SVDD242 pKa = 3.32GGFIAGTVNTEE253 pKa = 3.59LWTLAADD260 pKa = 3.75YY261 pKa = 11.65AMEE264 pKa = 4.19NTNFGISYY272 pKa = 9.4MDD274 pKa = 3.4YY275 pKa = 10.99QEE277 pKa = 4.29SQKK280 pKa = 11.46SNDD283 pKa = 2.75IDD285 pKa = 4.0ADD287 pKa = 3.32EE288 pKa = 4.32WGVYY292 pKa = 9.34VSHH295 pKa = 7.13KK296 pKa = 10.25LSNKK300 pKa = 9.87ASVKK304 pKa = 10.14AQYY307 pKa = 10.12TSADD311 pKa = 3.25VDD313 pKa = 3.46NTDD316 pKa = 3.46IVGEE320 pKa = 4.13DD321 pKa = 3.75VFVVGYY327 pKa = 9.93NVSFF331 pKa = 4.04
Molecular weight: 35.07 kDa
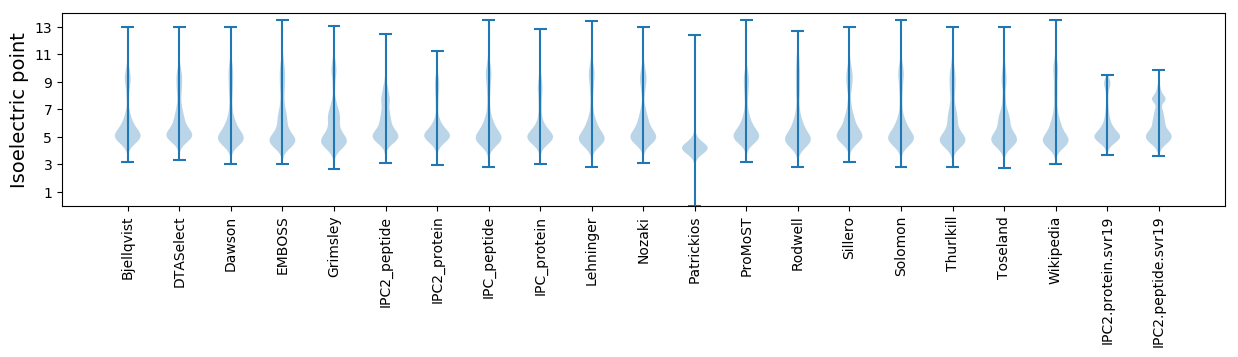
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H6DHK0|A0A1H6DHK0_9GAMM Uncharacterized protein OS=Marinobacterium lutimaris OX=568106 GN=SAMN05444390_106107 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.37RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.45NGRR28 pKa = 11.84QVLNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.7GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.37RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.45NGRR28 pKa = 11.84QVLNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.7GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1652987 |
27 |
10227 |
326.6 |
35.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.048 ± 0.044 | 1.012 ± 0.015 |
5.676 ± 0.047 | 6.616 ± 0.035 |
3.686 ± 0.023 | 7.771 ± 0.05 |
2.085 ± 0.02 | 5.452 ± 0.026 |
3.653 ± 0.032 | 11.18 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.532 ± 0.023 | 3.266 ± 0.024 |
4.506 ± 0.029 | 4.247 ± 0.024 |
6.016 ± 0.04 | 6.333 ± 0.032 |
4.984 ± 0.039 | 6.943 ± 0.032 |
1.316 ± 0.015 | 2.678 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
