
TM7 phylum sp. oral taxon 351
Taxonomy: cellular organisms; Bacteria; Bacteria incertae sedis; Bacteria candidate phyla; Candidatus Saccharibacteria
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
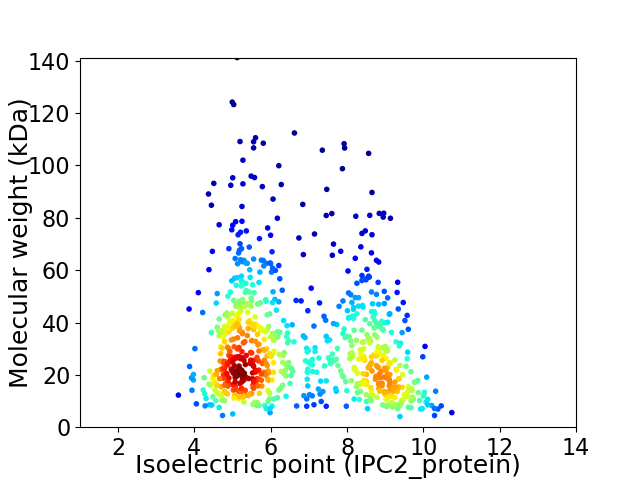
Virtual 2D-PAGE plot for 797 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A563C6B8|A0A563C6B8_9BACT Uncharacterized protein OS=TM7 phylum sp. oral taxon 351 OX=713053 GN=EUA77_03720 PE=4 SV=1
MM1 pKa = 7.7FEE3 pKa = 4.47EE4 pKa = 4.63YY5 pKa = 11.07NNGVPDD11 pKa = 4.21RR12 pKa = 11.84LRR14 pKa = 11.84QVTAEE19 pKa = 4.15TMSPEE24 pKa = 3.95ALYY27 pKa = 10.1TLDD30 pKa = 3.73TYY32 pKa = 11.83SDD34 pKa = 3.78VSPQDD39 pKa = 3.13VIYY42 pKa = 10.24IDD44 pKa = 3.65EE45 pKa = 4.33SRR47 pKa = 11.84AEE49 pKa = 3.99GSGRR53 pKa = 11.84LIISKK58 pKa = 9.42GQSVKK63 pKa = 10.76LLEE66 pKa = 4.52EE67 pKa = 5.2ADD69 pKa = 4.21LSTPDD74 pKa = 4.44LVILMSVWQRR84 pKa = 11.84NDD86 pKa = 3.0QGDD89 pKa = 3.84LCRR92 pKa = 11.84GIVADD97 pKa = 3.59CRR99 pKa = 11.84FYY101 pKa = 11.24SGCFDD106 pKa = 3.81AEE108 pKa = 4.31YY109 pKa = 10.8GDD111 pKa = 5.46EE112 pKa = 5.33EE113 pKa = 5.87IDD115 pKa = 3.96DD116 pKa = 4.55CDD118 pKa = 5.74GIDD121 pKa = 4.3DD122 pKa = 5.19EE123 pKa = 5.79IYY125 pKa = 11.18NKK127 pKa = 10.43DD128 pKa = 3.58DD129 pKa = 3.62EE130 pKa = 4.6QVISAVVDD138 pKa = 3.44YY139 pKa = 11.56GDD141 pKa = 4.33DD142 pKa = 3.74GKK144 pKa = 11.68VKK146 pKa = 10.03IWGDD150 pKa = 3.34PTFFDD155 pKa = 3.9AARR158 pKa = 11.84YY159 pKa = 7.2MAGLVDD165 pKa = 3.81NEE167 pKa = 4.52SIEE170 pKa = 4.21KK171 pKa = 10.96NEE173 pKa = 3.89VCASSHH179 pKa = 5.66EE180 pKa = 4.19LQDD183 pKa = 3.61CRR185 pKa = 11.84SYY187 pKa = 10.56QHH189 pKa = 6.35NVGASALPCSLIFLALASAII209 pKa = 3.95
MM1 pKa = 7.7FEE3 pKa = 4.47EE4 pKa = 4.63YY5 pKa = 11.07NNGVPDD11 pKa = 4.21RR12 pKa = 11.84LRR14 pKa = 11.84QVTAEE19 pKa = 4.15TMSPEE24 pKa = 3.95ALYY27 pKa = 10.1TLDD30 pKa = 3.73TYY32 pKa = 11.83SDD34 pKa = 3.78VSPQDD39 pKa = 3.13VIYY42 pKa = 10.24IDD44 pKa = 3.65EE45 pKa = 4.33SRR47 pKa = 11.84AEE49 pKa = 3.99GSGRR53 pKa = 11.84LIISKK58 pKa = 9.42GQSVKK63 pKa = 10.76LLEE66 pKa = 4.52EE67 pKa = 5.2ADD69 pKa = 4.21LSTPDD74 pKa = 4.44LVILMSVWQRR84 pKa = 11.84NDD86 pKa = 3.0QGDD89 pKa = 3.84LCRR92 pKa = 11.84GIVADD97 pKa = 3.59CRR99 pKa = 11.84FYY101 pKa = 11.24SGCFDD106 pKa = 3.81AEE108 pKa = 4.31YY109 pKa = 10.8GDD111 pKa = 5.46EE112 pKa = 5.33EE113 pKa = 5.87IDD115 pKa = 3.96DD116 pKa = 4.55CDD118 pKa = 5.74GIDD121 pKa = 4.3DD122 pKa = 5.19EE123 pKa = 5.79IYY125 pKa = 11.18NKK127 pKa = 10.43DD128 pKa = 3.58DD129 pKa = 3.62EE130 pKa = 4.6QVISAVVDD138 pKa = 3.44YY139 pKa = 11.56GDD141 pKa = 4.33DD142 pKa = 3.74GKK144 pKa = 11.68VKK146 pKa = 10.03IWGDD150 pKa = 3.34PTFFDD155 pKa = 3.9AARR158 pKa = 11.84YY159 pKa = 7.2MAGLVDD165 pKa = 3.81NEE167 pKa = 4.52SIEE170 pKa = 4.21KK171 pKa = 10.96NEE173 pKa = 3.89VCASSHH179 pKa = 5.66EE180 pKa = 4.19LQDD183 pKa = 3.61CRR185 pKa = 11.84SYY187 pKa = 10.56QHH189 pKa = 6.35NVGASALPCSLIFLALASAII209 pKa = 3.95
Molecular weight: 23.19 kDa
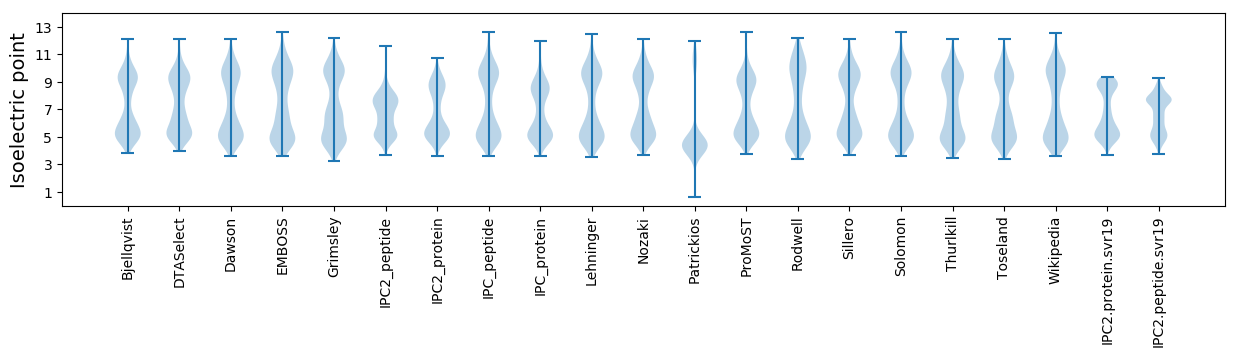
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A563CAN7|A0A563CAN7_9BACT Holliday junction ATP-dependent DNA helicase RuvB OS=TM7 phylum sp. oral taxon 351 OX=713053 GN=ruvB PE=3 SV=1
MM1 pKa = 7.29QDD3 pKa = 3.0EE4 pKa = 4.84SRR6 pKa = 11.84TSIRR10 pKa = 11.84ISQPSSARR18 pKa = 11.84TRR20 pKa = 11.84MAQSQASYY28 pKa = 10.69RR29 pKa = 11.84RR30 pKa = 11.84MDD32 pKa = 4.13FAPVQTVRR40 pKa = 11.84RR41 pKa = 11.84AAASIPTDD49 pKa = 3.32RR50 pKa = 11.84DD51 pKa = 3.44LEE53 pKa = 4.02RR54 pKa = 11.84HH55 pKa = 6.27RR56 pKa = 11.84IAKK59 pKa = 9.75QEE61 pKa = 3.89MQRR64 pKa = 11.84RR65 pKa = 11.84EE66 pKa = 4.49AIRR69 pKa = 11.84QQIEE73 pKa = 3.78EE74 pKa = 4.06QNRR77 pKa = 11.84LKK79 pKa = 10.88NLNQRR84 pKa = 11.84RR85 pKa = 11.84IEE87 pKa = 3.95AARR90 pKa = 11.84QEE92 pKa = 4.1LSEE95 pKa = 4.11YY96 pKa = 9.98QAAEE100 pKa = 4.1EE101 pKa = 3.98QRR103 pKa = 11.84TRR105 pKa = 11.84ALEE108 pKa = 3.91AEE110 pKa = 4.24RR111 pKa = 11.84QRR113 pKa = 11.84AIEE116 pKa = 4.16SQRR119 pKa = 11.84LAQRR123 pKa = 11.84EE124 pKa = 4.26ALEE127 pKa = 4.07KK128 pKa = 9.16MAARR132 pKa = 11.84RR133 pKa = 11.84RR134 pKa = 11.84AMEE137 pKa = 3.46QEE139 pKa = 3.3ARR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 11.84QIEE146 pKa = 3.95EE147 pKa = 3.46QDD149 pKa = 2.94RR150 pKa = 11.84KK151 pKa = 8.99MRR153 pKa = 11.84AMQEE157 pKa = 3.79MQRR160 pKa = 11.84QQEE163 pKa = 3.78ASKK166 pKa = 10.56INVSRR171 pKa = 11.84RR172 pKa = 11.84ISVAPVQTSPFKK184 pKa = 10.94KK185 pKa = 8.78VTNPAARR192 pKa = 11.84IQARR196 pKa = 11.84SVDD199 pKa = 3.92MQAKK203 pKa = 5.59TTISVSDD210 pKa = 3.82TQAKK214 pKa = 8.22PALRR218 pKa = 11.84SAGDD222 pKa = 3.74TKK224 pKa = 11.03VLPASRR230 pKa = 11.84IQVVASPNRR239 pKa = 11.84KK240 pKa = 8.7LFGRR244 pKa = 11.84IRR246 pKa = 11.84PAGPSRR252 pKa = 11.84QVPQDD257 pKa = 3.39EE258 pKa = 4.16QDD260 pKa = 3.84PLVQSARR267 pKa = 11.84NSHH270 pKa = 6.4IEE272 pKa = 3.77LSTSGASITSDD283 pKa = 3.09SDD285 pKa = 4.24LEE287 pKa = 4.38DD288 pKa = 3.34MLYY291 pKa = 10.87DD292 pKa = 3.64AFEE295 pKa = 5.67DD296 pKa = 4.08STSDD300 pKa = 3.19TPEE303 pKa = 3.99RR304 pKa = 11.84LSSKK308 pKa = 10.37SRR310 pKa = 11.84NQSQNSS316 pKa = 3.35
MM1 pKa = 7.29QDD3 pKa = 3.0EE4 pKa = 4.84SRR6 pKa = 11.84TSIRR10 pKa = 11.84ISQPSSARR18 pKa = 11.84TRR20 pKa = 11.84MAQSQASYY28 pKa = 10.69RR29 pKa = 11.84RR30 pKa = 11.84MDD32 pKa = 4.13FAPVQTVRR40 pKa = 11.84RR41 pKa = 11.84AAASIPTDD49 pKa = 3.32RR50 pKa = 11.84DD51 pKa = 3.44LEE53 pKa = 4.02RR54 pKa = 11.84HH55 pKa = 6.27RR56 pKa = 11.84IAKK59 pKa = 9.75QEE61 pKa = 3.89MQRR64 pKa = 11.84RR65 pKa = 11.84EE66 pKa = 4.49AIRR69 pKa = 11.84QQIEE73 pKa = 3.78EE74 pKa = 4.06QNRR77 pKa = 11.84LKK79 pKa = 10.88NLNQRR84 pKa = 11.84RR85 pKa = 11.84IEE87 pKa = 3.95AARR90 pKa = 11.84QEE92 pKa = 4.1LSEE95 pKa = 4.11YY96 pKa = 9.98QAAEE100 pKa = 4.1EE101 pKa = 3.98QRR103 pKa = 11.84TRR105 pKa = 11.84ALEE108 pKa = 3.91AEE110 pKa = 4.24RR111 pKa = 11.84QRR113 pKa = 11.84AIEE116 pKa = 4.16SQRR119 pKa = 11.84LAQRR123 pKa = 11.84EE124 pKa = 4.26ALEE127 pKa = 4.07KK128 pKa = 9.16MAARR132 pKa = 11.84RR133 pKa = 11.84RR134 pKa = 11.84AMEE137 pKa = 3.46QEE139 pKa = 3.3ARR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 11.84QIEE146 pKa = 3.95EE147 pKa = 3.46QDD149 pKa = 2.94RR150 pKa = 11.84KK151 pKa = 8.99MRR153 pKa = 11.84AMQEE157 pKa = 3.79MQRR160 pKa = 11.84QQEE163 pKa = 3.78ASKK166 pKa = 10.56INVSRR171 pKa = 11.84RR172 pKa = 11.84ISVAPVQTSPFKK184 pKa = 10.94KK185 pKa = 8.78VTNPAARR192 pKa = 11.84IQARR196 pKa = 11.84SVDD199 pKa = 3.92MQAKK203 pKa = 5.59TTISVSDD210 pKa = 3.82TQAKK214 pKa = 8.22PALRR218 pKa = 11.84SAGDD222 pKa = 3.74TKK224 pKa = 11.03VLPASRR230 pKa = 11.84IQVVASPNRR239 pKa = 11.84KK240 pKa = 8.7LFGRR244 pKa = 11.84IRR246 pKa = 11.84PAGPSRR252 pKa = 11.84QVPQDD257 pKa = 3.39EE258 pKa = 4.16QDD260 pKa = 3.84PLVQSARR267 pKa = 11.84NSHH270 pKa = 6.4IEE272 pKa = 3.77LSTSGASITSDD283 pKa = 3.09SDD285 pKa = 4.24LEE287 pKa = 4.38DD288 pKa = 3.34MLYY291 pKa = 10.87DD292 pKa = 3.64AFEE295 pKa = 5.67DD296 pKa = 4.08STSDD300 pKa = 3.19TPEE303 pKa = 3.99RR304 pKa = 11.84LSSKK308 pKa = 10.37SRR310 pKa = 11.84NQSQNSS316 pKa = 3.35
Molecular weight: 36.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
235844 |
38 |
1280 |
295.9 |
33.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.545 ± 0.08 | 0.639 ± 0.024 |
5.837 ± 0.069 | 6.884 ± 0.099 |
4.046 ± 0.061 | 6.418 ± 0.083 |
1.749 ± 0.038 | 7.543 ± 0.08 |
7.425 ± 0.088 | 9.246 ± 0.077 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.369 ± 0.04 | 4.987 ± 0.08 |
3.641 ± 0.055 | 3.685 ± 0.061 |
4.765 ± 0.071 | 6.663 ± 0.082 |
5.691 ± 0.078 | 6.643 ± 0.069 |
0.896 ± 0.032 | 3.327 ± 0.051 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
