
Oceanibaculum indicum P24
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Thalassobaculaceae; Oceanibaculum; Oceanibaculum indicum
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
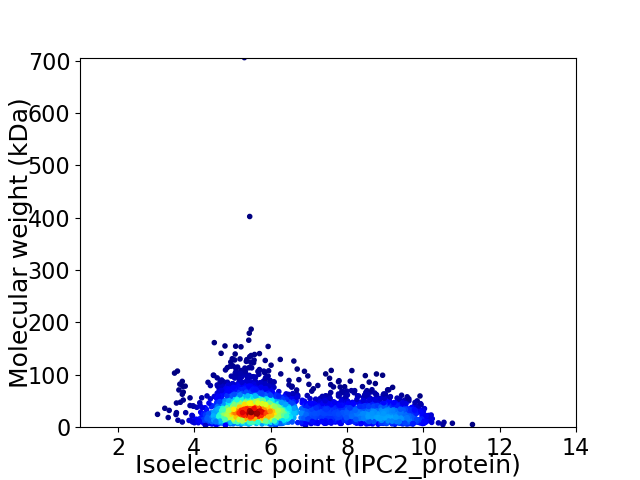
Virtual 2D-PAGE plot for 3754 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K2KHU1|K2KHU1_9PROT Ribokinase-like domain-containing protein OS=Oceanibaculum indicum P24 OX=1207063 GN=P24_06671 PE=4 SV=1
MM1 pKa = 6.69TTTTAVTASGTSYY14 pKa = 10.7IDD16 pKa = 3.89SLLHH20 pKa = 4.8GQKK23 pKa = 9.5WDD25 pKa = 3.69GTTLTYY31 pKa = 10.87AFPDD35 pKa = 3.13AAANVGYY42 pKa = 10.14SIDD45 pKa = 3.62GAYY48 pKa = 7.82FTALSGAEE56 pKa = 3.75QTAFEE61 pKa = 4.71AVLQAWARR69 pKa = 11.84VSGLTFTEE77 pKa = 4.41VAAADD82 pKa = 3.78AATADD87 pKa = 3.52ISVYY91 pKa = 9.4WYY93 pKa = 10.26RR94 pKa = 11.84SPDD97 pKa = 3.31NLTARR102 pKa = 11.84VVTFPDD108 pKa = 3.79GTVEE112 pKa = 4.83GGDD115 pKa = 3.33IQLGSGINGGDD126 pKa = 3.35LSSPGSYY133 pKa = 9.95SHH135 pKa = 7.51FIAMHH140 pKa = 5.78EE141 pKa = 4.28LGHH144 pKa = 6.86ALGLKK149 pKa = 9.75HH150 pKa = 5.66PHH152 pKa = 5.47EE153 pKa = 4.84VEE155 pKa = 4.08GSFPADD161 pKa = 2.52TGMSVEE167 pKa = 4.48LSVMSYY173 pKa = 10.83VSFTGGSVSSYY184 pKa = 10.87SIADD188 pKa = 3.41GSYY191 pKa = 8.3PTAPMLNDD199 pKa = 3.28IAAIQYY205 pKa = 10.58LYY207 pKa = 11.13GVNTNALTANLGDD220 pKa = 3.4TTYY223 pKa = 10.51TYY225 pKa = 11.31DD226 pKa = 3.76PGAAIIFQARR236 pKa = 11.84WDD238 pKa = 3.63GGGYY242 pKa = 8.4DD243 pKa = 3.57TFNFASYY250 pKa = 8.4GTDD253 pKa = 2.87LSVDD257 pKa = 3.9LRR259 pKa = 11.84PGQWIDD265 pKa = 4.11LGGQYY270 pKa = 11.11AVLDD274 pKa = 3.76TGDD277 pKa = 3.64INVTPLGNIAIPYY290 pKa = 8.17LHH292 pKa = 7.22EE293 pKa = 5.13GNTAYY298 pKa = 10.82LIEE301 pKa = 4.26AAYY304 pKa = 10.07GGSGNDD310 pKa = 3.52LIVGNEE316 pKa = 3.82ANNLLRR322 pKa = 11.84GNGGNDD328 pKa = 3.49TLRR331 pKa = 11.84GGAGTDD337 pKa = 3.48TLVGSAGQDD346 pKa = 2.96TFDD349 pKa = 4.08FTDD352 pKa = 4.42GAASSNTVSDD362 pKa = 4.22FTADD366 pKa = 3.44DD367 pKa = 4.36TIVLRR372 pKa = 11.84DD373 pKa = 3.67AVVGGIVRR381 pKa = 11.84GNDD384 pKa = 3.25GATLAQGEE392 pKa = 4.91VSFAWNGVYY401 pKa = 10.34NVLYY405 pKa = 10.75VGLDD409 pKa = 3.26ATAGSDD415 pKa = 3.85LQIVLANSTVFDD427 pKa = 4.03NLSLSGNTISFIADD441 pKa = 3.52TVAPEE446 pKa = 4.17FLSGTGPADD455 pKa = 3.32NAVGVARR462 pKa = 11.84DD463 pKa = 3.82ATPTLIFNEE472 pKa = 4.36EE473 pKa = 3.77VSAGTGEE480 pKa = 4.01FIIYY484 pKa = 10.36NVTNPSVVKK493 pKa = 9.67TIAANSAAVTGWGTYY508 pKa = 10.02RR509 pKa = 11.84LTIDD513 pKa = 4.12IDD515 pKa = 4.06PDD517 pKa = 3.63GLTPLPYY524 pKa = 10.01GAEE527 pKa = 3.79IAIMWDD533 pKa = 3.16EE534 pKa = 4.14SAVRR538 pKa = 11.84DD539 pKa = 3.99LAGNPLAEE547 pKa = 4.39NTSEE551 pKa = 4.04TLYY554 pKa = 11.36SFIANVGPTATASAANVAHH573 pKa = 6.55TAAGQTSYY581 pKa = 11.34SFTVTYY587 pKa = 10.91SDD589 pKa = 4.25ADD591 pKa = 3.62GTIDD595 pKa = 4.67DD596 pKa = 4.66TSIDD600 pKa = 4.02TNDD603 pKa = 3.55VTVTAPDD610 pKa = 3.65TSTLTVTGAVWNGDD624 pKa = 3.8TNTATYY630 pKa = 8.85TVEE633 pKa = 4.13VPGGNGWEE641 pKa = 3.99AALEE645 pKa = 4.07GTYY648 pKa = 10.21TIGLVEE654 pKa = 5.26GEE656 pKa = 4.29VTDD659 pKa = 3.53NHH661 pKa = 6.68GDD663 pKa = 3.74GVAGNANADD672 pKa = 3.84SFTVDD677 pKa = 3.34LTVPPTPDD685 pKa = 4.0PGPGPSLPPPTDD697 pKa = 3.21TSVGGVDD704 pKa = 2.6IATRR708 pKa = 11.84YY709 pKa = 8.18STDD712 pKa = 2.98ASDD715 pKa = 3.66RR716 pKa = 11.84QIEE719 pKa = 4.57TVTITPNGTQQTTQGVPLAGPASNPVLSGTLPSTVSVTVTRR760 pKa = 11.84PSAPISSTALVNSLLAEE777 pKa = 4.27VPFGIVLPDD786 pKa = 4.15DD787 pKa = 4.1PSLPVLAALRR797 pKa = 11.84PNVPITARR805 pKa = 11.84TITMTPYY812 pKa = 9.85GTDD815 pKa = 3.52DD816 pKa = 4.26LLEE819 pKa = 4.45AMIIEE824 pKa = 5.03GSPDD828 pKa = 2.96SGQASGIVLDD838 pKa = 4.73ARR840 pKa = 11.84NLPSGAPIQLNNVDD854 pKa = 3.61VAIISGDD861 pKa = 3.7VVVSGGAGPSIVLGDD876 pKa = 4.08AGSQNIVLGEE886 pKa = 4.09GTDD889 pKa = 3.93YY890 pKa = 11.63LHH892 pKa = 7.22GGAGNDD898 pKa = 4.07TIGSATGSDD907 pKa = 3.27TLLGGEE913 pKa = 4.67GDD915 pKa = 3.91DD916 pKa = 4.77SVFGGMNGDD925 pKa = 4.04SIEE928 pKa = 4.34GGEE931 pKa = 4.44GNDD934 pKa = 3.26ILRR937 pKa = 11.84GGKK940 pKa = 9.36GHH942 pKa = 7.79DD943 pKa = 4.12SLDD946 pKa = 3.55GGAGDD951 pKa = 5.55DD952 pKa = 3.44ILYY955 pKa = 10.82SGFGDD960 pKa = 3.64DD961 pKa = 4.01TLTGGEE967 pKa = 4.21GADD970 pKa = 3.41LFVLRR975 pKa = 11.84GFDD978 pKa = 3.68ANFAGAVLKK987 pKa = 10.12ATVTDD992 pKa = 4.89FAQGTDD998 pKa = 2.92RR999 pKa = 11.84FAVEE1003 pKa = 4.27NVSVAALEE1011 pKa = 4.24AALALQSVTEE1021 pKa = 4.15AGVVIEE1027 pKa = 4.27VAGATLTFIGISQLTAADD1045 pKa = 3.77IDD1047 pKa = 3.63QSFYY1051 pKa = 11.76AA1052 pKa = 4.75
MM1 pKa = 6.69TTTTAVTASGTSYY14 pKa = 10.7IDD16 pKa = 3.89SLLHH20 pKa = 4.8GQKK23 pKa = 9.5WDD25 pKa = 3.69GTTLTYY31 pKa = 10.87AFPDD35 pKa = 3.13AAANVGYY42 pKa = 10.14SIDD45 pKa = 3.62GAYY48 pKa = 7.82FTALSGAEE56 pKa = 3.75QTAFEE61 pKa = 4.71AVLQAWARR69 pKa = 11.84VSGLTFTEE77 pKa = 4.41VAAADD82 pKa = 3.78AATADD87 pKa = 3.52ISVYY91 pKa = 9.4WYY93 pKa = 10.26RR94 pKa = 11.84SPDD97 pKa = 3.31NLTARR102 pKa = 11.84VVTFPDD108 pKa = 3.79GTVEE112 pKa = 4.83GGDD115 pKa = 3.33IQLGSGINGGDD126 pKa = 3.35LSSPGSYY133 pKa = 9.95SHH135 pKa = 7.51FIAMHH140 pKa = 5.78EE141 pKa = 4.28LGHH144 pKa = 6.86ALGLKK149 pKa = 9.75HH150 pKa = 5.66PHH152 pKa = 5.47EE153 pKa = 4.84VEE155 pKa = 4.08GSFPADD161 pKa = 2.52TGMSVEE167 pKa = 4.48LSVMSYY173 pKa = 10.83VSFTGGSVSSYY184 pKa = 10.87SIADD188 pKa = 3.41GSYY191 pKa = 8.3PTAPMLNDD199 pKa = 3.28IAAIQYY205 pKa = 10.58LYY207 pKa = 11.13GVNTNALTANLGDD220 pKa = 3.4TTYY223 pKa = 10.51TYY225 pKa = 11.31DD226 pKa = 3.76PGAAIIFQARR236 pKa = 11.84WDD238 pKa = 3.63GGGYY242 pKa = 8.4DD243 pKa = 3.57TFNFASYY250 pKa = 8.4GTDD253 pKa = 2.87LSVDD257 pKa = 3.9LRR259 pKa = 11.84PGQWIDD265 pKa = 4.11LGGQYY270 pKa = 11.11AVLDD274 pKa = 3.76TGDD277 pKa = 3.64INVTPLGNIAIPYY290 pKa = 8.17LHH292 pKa = 7.22EE293 pKa = 5.13GNTAYY298 pKa = 10.82LIEE301 pKa = 4.26AAYY304 pKa = 10.07GGSGNDD310 pKa = 3.52LIVGNEE316 pKa = 3.82ANNLLRR322 pKa = 11.84GNGGNDD328 pKa = 3.49TLRR331 pKa = 11.84GGAGTDD337 pKa = 3.48TLVGSAGQDD346 pKa = 2.96TFDD349 pKa = 4.08FTDD352 pKa = 4.42GAASSNTVSDD362 pKa = 4.22FTADD366 pKa = 3.44DD367 pKa = 4.36TIVLRR372 pKa = 11.84DD373 pKa = 3.67AVVGGIVRR381 pKa = 11.84GNDD384 pKa = 3.25GATLAQGEE392 pKa = 4.91VSFAWNGVYY401 pKa = 10.34NVLYY405 pKa = 10.75VGLDD409 pKa = 3.26ATAGSDD415 pKa = 3.85LQIVLANSTVFDD427 pKa = 4.03NLSLSGNTISFIADD441 pKa = 3.52TVAPEE446 pKa = 4.17FLSGTGPADD455 pKa = 3.32NAVGVARR462 pKa = 11.84DD463 pKa = 3.82ATPTLIFNEE472 pKa = 4.36EE473 pKa = 3.77VSAGTGEE480 pKa = 4.01FIIYY484 pKa = 10.36NVTNPSVVKK493 pKa = 9.67TIAANSAAVTGWGTYY508 pKa = 10.02RR509 pKa = 11.84LTIDD513 pKa = 4.12IDD515 pKa = 4.06PDD517 pKa = 3.63GLTPLPYY524 pKa = 10.01GAEE527 pKa = 3.79IAIMWDD533 pKa = 3.16EE534 pKa = 4.14SAVRR538 pKa = 11.84DD539 pKa = 3.99LAGNPLAEE547 pKa = 4.39NTSEE551 pKa = 4.04TLYY554 pKa = 11.36SFIANVGPTATASAANVAHH573 pKa = 6.55TAAGQTSYY581 pKa = 11.34SFTVTYY587 pKa = 10.91SDD589 pKa = 4.25ADD591 pKa = 3.62GTIDD595 pKa = 4.67DD596 pKa = 4.66TSIDD600 pKa = 4.02TNDD603 pKa = 3.55VTVTAPDD610 pKa = 3.65TSTLTVTGAVWNGDD624 pKa = 3.8TNTATYY630 pKa = 8.85TVEE633 pKa = 4.13VPGGNGWEE641 pKa = 3.99AALEE645 pKa = 4.07GTYY648 pKa = 10.21TIGLVEE654 pKa = 5.26GEE656 pKa = 4.29VTDD659 pKa = 3.53NHH661 pKa = 6.68GDD663 pKa = 3.74GVAGNANADD672 pKa = 3.84SFTVDD677 pKa = 3.34LTVPPTPDD685 pKa = 4.0PGPGPSLPPPTDD697 pKa = 3.21TSVGGVDD704 pKa = 2.6IATRR708 pKa = 11.84YY709 pKa = 8.18STDD712 pKa = 2.98ASDD715 pKa = 3.66RR716 pKa = 11.84QIEE719 pKa = 4.57TVTITPNGTQQTTQGVPLAGPASNPVLSGTLPSTVSVTVTRR760 pKa = 11.84PSAPISSTALVNSLLAEE777 pKa = 4.27VPFGIVLPDD786 pKa = 4.15DD787 pKa = 4.1PSLPVLAALRR797 pKa = 11.84PNVPITARR805 pKa = 11.84TITMTPYY812 pKa = 9.85GTDD815 pKa = 3.52DD816 pKa = 4.26LLEE819 pKa = 4.45AMIIEE824 pKa = 5.03GSPDD828 pKa = 2.96SGQASGIVLDD838 pKa = 4.73ARR840 pKa = 11.84NLPSGAPIQLNNVDD854 pKa = 3.61VAIISGDD861 pKa = 3.7VVVSGGAGPSIVLGDD876 pKa = 4.08AGSQNIVLGEE886 pKa = 4.09GTDD889 pKa = 3.93YY890 pKa = 11.63LHH892 pKa = 7.22GGAGNDD898 pKa = 4.07TIGSATGSDD907 pKa = 3.27TLLGGEE913 pKa = 4.67GDD915 pKa = 3.91DD916 pKa = 4.77SVFGGMNGDD925 pKa = 4.04SIEE928 pKa = 4.34GGEE931 pKa = 4.44GNDD934 pKa = 3.26ILRR937 pKa = 11.84GGKK940 pKa = 9.36GHH942 pKa = 7.79DD943 pKa = 4.12SLDD946 pKa = 3.55GGAGDD951 pKa = 5.55DD952 pKa = 3.44ILYY955 pKa = 10.82SGFGDD960 pKa = 3.64DD961 pKa = 4.01TLTGGEE967 pKa = 4.21GADD970 pKa = 3.41LFVLRR975 pKa = 11.84GFDD978 pKa = 3.68ANFAGAVLKK987 pKa = 10.12ATVTDD992 pKa = 4.89FAQGTDD998 pKa = 2.92RR999 pKa = 11.84FAVEE1003 pKa = 4.27NVSVAALEE1011 pKa = 4.24AALALQSVTEE1021 pKa = 4.15AGVVIEE1027 pKa = 4.27VAGATLTFIGISQLTAADD1045 pKa = 3.77IDD1047 pKa = 3.63QSFYY1051 pKa = 11.76AA1052 pKa = 4.75
Molecular weight: 107.11 kDa
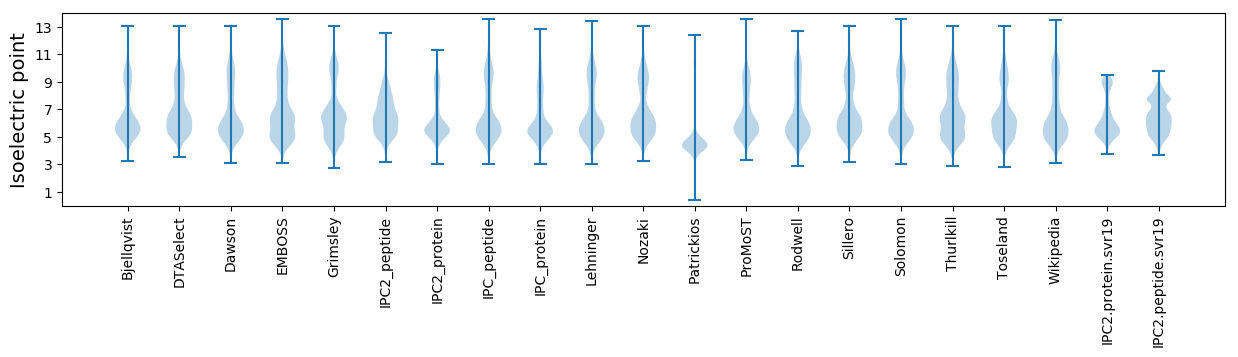
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K2KLP3|K2KLP3_9PROT YlxR domain-containing protein OS=Oceanibaculum indicum P24 OX=1207063 GN=P24_02326 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 8.7IVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.49GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.32VIANRR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.9RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 8.7IVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.49GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.32VIANRR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.9RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1192065 |
27 |
6700 |
317.5 |
34.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.631 ± 0.062 | 0.837 ± 0.012 |
5.65 ± 0.036 | 5.898 ± 0.039 |
3.546 ± 0.027 | 8.851 ± 0.04 |
1.968 ± 0.018 | 5.051 ± 0.033 |
3.25 ± 0.039 | 10.715 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.612 ± 0.023 | 2.417 ± 0.022 |
5.328 ± 0.037 | 3.197 ± 0.023 |
7.12 ± 0.041 | 4.956 ± 0.021 |
5.17 ± 0.028 | 7.209 ± 0.036 |
1.288 ± 0.016 | 2.306 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
