
Zunongwangia atlantica 22II14-10F7
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Zunongwangia; Zunongwangia atlantica
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
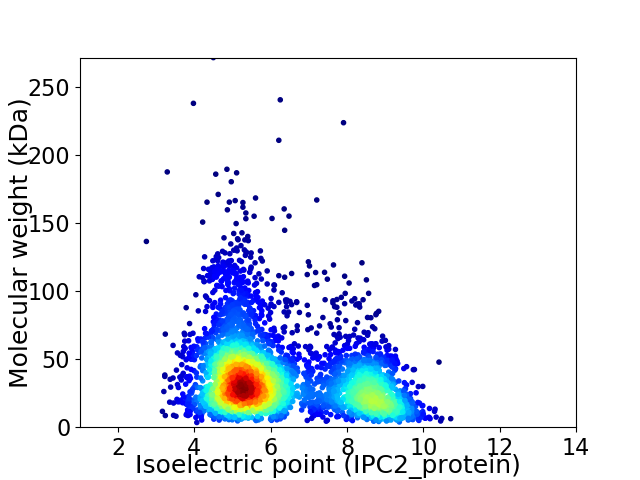
Virtual 2D-PAGE plot for 4100 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y1T682|A0A1Y1T682_9FLAO DUF3817 domain-containing protein OS=Zunongwangia atlantica 22II14-10F7 OX=1185767 GN=IIF7_05872 PE=4 SV=1
MM1 pKa = 7.47KK2 pKa = 10.34KK3 pKa = 10.74SNFNQLFKK11 pKa = 10.84PVIFTLGILLFSINLEE27 pKa = 4.08AQCPIGNVTINSQSEE42 pKa = 4.37LIQFQNNYY50 pKa = 8.71PNCTIINANLSISSLVTDD68 pKa = 4.46LTPLQNIEE76 pKa = 4.45EE77 pKa = 4.36IKK79 pKa = 11.08GDD81 pKa = 3.7LTISNTNIEE90 pKa = 4.55DD91 pKa = 3.73LTTEE95 pKa = 4.47LSISKK100 pKa = 10.42INGDD104 pKa = 4.31LNIINNNLLRR114 pKa = 11.84HH115 pKa = 5.3VNSFNEE121 pKa = 4.06LTEE124 pKa = 3.89VSNINIDD131 pKa = 3.9RR132 pKa = 11.84ISGYY136 pKa = 10.43FDD138 pKa = 3.62GFNKK142 pKa = 10.32LEE144 pKa = 4.08KK145 pKa = 11.09VNTIDD150 pKa = 5.09LNNLGMSLDD159 pKa = 3.53AFQNLKK165 pKa = 10.35FVEE168 pKa = 4.56EE169 pKa = 4.14SLIMDD174 pKa = 4.85DD175 pKa = 5.07LGLALRR181 pKa = 11.84IYY183 pKa = 10.11QAFNKK188 pKa = 9.7VEE190 pKa = 4.18YY191 pKa = 10.02IKK193 pKa = 10.96GSLVIRR199 pKa = 11.84DD200 pKa = 3.89YY201 pKa = 11.33PHH203 pKa = 7.18GIGLFSEE210 pKa = 4.83LKK212 pKa = 10.94YY213 pKa = 10.59IGNDD217 pKa = 3.08LFINRR222 pKa = 11.84TGINEE227 pKa = 3.68IEE229 pKa = 4.38GFNKK233 pKa = 10.4LEE235 pKa = 4.67SIGSLGGGDD244 pKa = 4.07FEE246 pKa = 4.4ITEE249 pKa = 4.34SLEE252 pKa = 3.95LTVFTGFNNLQRR264 pKa = 11.84VYY266 pKa = 10.83GNMVITGNPEE276 pKa = 3.6LPEE279 pKa = 4.21ISGFYY284 pKa = 10.29NLEE287 pKa = 4.29KK288 pKa = 9.67IDD290 pKa = 3.83RR291 pKa = 11.84ALIIQNNNRR300 pKa = 11.84LAALTGLEE308 pKa = 3.93NLNAVSNNFGNLGLIVSNNQEE329 pKa = 4.16LTDD332 pKa = 4.42CSAICNLLANNRR344 pKa = 11.84VYY346 pKa = 11.47DD347 pKa = 4.45DD348 pKa = 4.78VEE350 pKa = 4.0ILNNPSEE357 pKa = 4.31CSTRR361 pKa = 11.84QEE363 pKa = 4.18IEE365 pKa = 4.6SNCLPDD371 pKa = 4.43FDD373 pKa = 5.87NDD375 pKa = 5.46GIPDD379 pKa = 5.66DD380 pKa = 5.49IDD382 pKa = 6.38LDD384 pKa = 4.39DD385 pKa = 6.24DD386 pKa = 4.26NDD388 pKa = 5.07GIPDD392 pKa = 4.22IIEE395 pKa = 4.54DD396 pKa = 3.79NGILDD401 pKa = 4.3RR402 pKa = 11.84DD403 pKa = 3.83SDD405 pKa = 4.26NDD407 pKa = 3.58GSPDD411 pKa = 4.42KK412 pKa = 10.95RR413 pKa = 11.84DD414 pKa = 3.51LDD416 pKa = 4.05SDD418 pKa = 3.78NDD420 pKa = 3.58GCYY423 pKa = 10.52DD424 pKa = 3.49VLEE427 pKa = 5.0AGFNDD432 pKa = 4.64DD433 pKa = 5.43DD434 pKa = 6.0DD435 pKa = 5.55NGTLGNSPDD444 pKa = 3.82TVDD447 pKa = 3.97EE448 pKa = 5.04DD449 pKa = 3.95GLIINEE455 pKa = 4.29TTGYY459 pKa = 6.34TTPLDD464 pKa = 3.75EE465 pKa = 5.69DD466 pKa = 3.69NNGIYY471 pKa = 10.42DD472 pKa = 3.71FQEE475 pKa = 4.22AFTLSAGNDD484 pKa = 3.27ASINLCQEE492 pKa = 3.72GNAIDD497 pKa = 3.97LFNYY501 pKa = 9.82LGKK504 pKa = 10.14DD505 pKa = 3.36AQRR508 pKa = 11.84GGTWSPNLASNSSVFDD524 pKa = 3.95PSLDD528 pKa = 3.58SEE530 pKa = 4.85GIYY533 pKa = 10.3AYY535 pKa = 9.05TVNNYY540 pKa = 9.96CISQTAIIEE549 pKa = 4.54INFEE553 pKa = 4.08PQPEE557 pKa = 4.37PGLDD561 pKa = 3.54TNISICQYY569 pKa = 9.87EE570 pKa = 4.2EE571 pKa = 3.67AFRR574 pKa = 11.84LVDD577 pKa = 3.79YY578 pKa = 11.6LEE580 pKa = 5.06GNPDD584 pKa = 3.56SNGYY588 pKa = 8.71WEE590 pKa = 4.68PSLPNGIFDD599 pKa = 3.86PQTNEE604 pKa = 3.56EE605 pKa = 4.1GTYY608 pKa = 9.52TYY610 pKa = 10.32HH611 pKa = 7.49ISNEE615 pKa = 3.89NCEE618 pKa = 4.25VLTSEE623 pKa = 5.42IIISFLDD630 pKa = 3.32RR631 pKa = 11.84TEE633 pKa = 4.42TISYY637 pKa = 9.36EE638 pKa = 3.93YY639 pKa = 10.04STALNNDD646 pKa = 2.42GSYY649 pKa = 11.31NIFFEE654 pKa = 4.49TANEE658 pKa = 4.01SGYY661 pKa = 9.03TFSLDD666 pKa = 4.89DD667 pKa = 4.18MTDD670 pKa = 2.88QSNGYY675 pKa = 9.21FYY677 pKa = 10.81NISPGIHH684 pKa = 5.65QIKK687 pKa = 8.0ITEE690 pKa = 4.08INGCGIVNEE699 pKa = 4.24QIALIGFPKK708 pKa = 10.31YY709 pKa = 10.6FSPNSDD715 pKa = 2.71GMNDD719 pKa = 2.9FWQPLGIIDD728 pKa = 5.29NEE730 pKa = 4.35VQTYY734 pKa = 9.97IYY736 pKa = 10.65DD737 pKa = 3.4RR738 pKa = 11.84YY739 pKa = 9.44GTLLRR744 pKa = 11.84KK745 pKa = 9.57LQGDD749 pKa = 3.93EE750 pKa = 4.41HH751 pKa = 6.95WDD753 pKa = 3.13GSYY756 pKa = 10.63NGKK759 pKa = 9.42PMPEE763 pKa = 3.5DD764 pKa = 4.69DD765 pKa = 4.11YY766 pKa = 11.47WFKK769 pKa = 11.54ALIGQEE775 pKa = 4.32SEE777 pKa = 4.4QKK779 pKa = 10.8GHH781 pKa = 6.76FSLIRR786 pKa = 4.08
MM1 pKa = 7.47KK2 pKa = 10.34KK3 pKa = 10.74SNFNQLFKK11 pKa = 10.84PVIFTLGILLFSINLEE27 pKa = 4.08AQCPIGNVTINSQSEE42 pKa = 4.37LIQFQNNYY50 pKa = 8.71PNCTIINANLSISSLVTDD68 pKa = 4.46LTPLQNIEE76 pKa = 4.45EE77 pKa = 4.36IKK79 pKa = 11.08GDD81 pKa = 3.7LTISNTNIEE90 pKa = 4.55DD91 pKa = 3.73LTTEE95 pKa = 4.47LSISKK100 pKa = 10.42INGDD104 pKa = 4.31LNIINNNLLRR114 pKa = 11.84HH115 pKa = 5.3VNSFNEE121 pKa = 4.06LTEE124 pKa = 3.89VSNINIDD131 pKa = 3.9RR132 pKa = 11.84ISGYY136 pKa = 10.43FDD138 pKa = 3.62GFNKK142 pKa = 10.32LEE144 pKa = 4.08KK145 pKa = 11.09VNTIDD150 pKa = 5.09LNNLGMSLDD159 pKa = 3.53AFQNLKK165 pKa = 10.35FVEE168 pKa = 4.56EE169 pKa = 4.14SLIMDD174 pKa = 4.85DD175 pKa = 5.07LGLALRR181 pKa = 11.84IYY183 pKa = 10.11QAFNKK188 pKa = 9.7VEE190 pKa = 4.18YY191 pKa = 10.02IKK193 pKa = 10.96GSLVIRR199 pKa = 11.84DD200 pKa = 3.89YY201 pKa = 11.33PHH203 pKa = 7.18GIGLFSEE210 pKa = 4.83LKK212 pKa = 10.94YY213 pKa = 10.59IGNDD217 pKa = 3.08LFINRR222 pKa = 11.84TGINEE227 pKa = 3.68IEE229 pKa = 4.38GFNKK233 pKa = 10.4LEE235 pKa = 4.67SIGSLGGGDD244 pKa = 4.07FEE246 pKa = 4.4ITEE249 pKa = 4.34SLEE252 pKa = 3.95LTVFTGFNNLQRR264 pKa = 11.84VYY266 pKa = 10.83GNMVITGNPEE276 pKa = 3.6LPEE279 pKa = 4.21ISGFYY284 pKa = 10.29NLEE287 pKa = 4.29KK288 pKa = 9.67IDD290 pKa = 3.83RR291 pKa = 11.84ALIIQNNNRR300 pKa = 11.84LAALTGLEE308 pKa = 3.93NLNAVSNNFGNLGLIVSNNQEE329 pKa = 4.16LTDD332 pKa = 4.42CSAICNLLANNRR344 pKa = 11.84VYY346 pKa = 11.47DD347 pKa = 4.45DD348 pKa = 4.78VEE350 pKa = 4.0ILNNPSEE357 pKa = 4.31CSTRR361 pKa = 11.84QEE363 pKa = 4.18IEE365 pKa = 4.6SNCLPDD371 pKa = 4.43FDD373 pKa = 5.87NDD375 pKa = 5.46GIPDD379 pKa = 5.66DD380 pKa = 5.49IDD382 pKa = 6.38LDD384 pKa = 4.39DD385 pKa = 6.24DD386 pKa = 4.26NDD388 pKa = 5.07GIPDD392 pKa = 4.22IIEE395 pKa = 4.54DD396 pKa = 3.79NGILDD401 pKa = 4.3RR402 pKa = 11.84DD403 pKa = 3.83SDD405 pKa = 4.26NDD407 pKa = 3.58GSPDD411 pKa = 4.42KK412 pKa = 10.95RR413 pKa = 11.84DD414 pKa = 3.51LDD416 pKa = 4.05SDD418 pKa = 3.78NDD420 pKa = 3.58GCYY423 pKa = 10.52DD424 pKa = 3.49VLEE427 pKa = 5.0AGFNDD432 pKa = 4.64DD433 pKa = 5.43DD434 pKa = 6.0DD435 pKa = 5.55NGTLGNSPDD444 pKa = 3.82TVDD447 pKa = 3.97EE448 pKa = 5.04DD449 pKa = 3.95GLIINEE455 pKa = 4.29TTGYY459 pKa = 6.34TTPLDD464 pKa = 3.75EE465 pKa = 5.69DD466 pKa = 3.69NNGIYY471 pKa = 10.42DD472 pKa = 3.71FQEE475 pKa = 4.22AFTLSAGNDD484 pKa = 3.27ASINLCQEE492 pKa = 3.72GNAIDD497 pKa = 3.97LFNYY501 pKa = 9.82LGKK504 pKa = 10.14DD505 pKa = 3.36AQRR508 pKa = 11.84GGTWSPNLASNSSVFDD524 pKa = 3.95PSLDD528 pKa = 3.58SEE530 pKa = 4.85GIYY533 pKa = 10.3AYY535 pKa = 9.05TVNNYY540 pKa = 9.96CISQTAIIEE549 pKa = 4.54INFEE553 pKa = 4.08PQPEE557 pKa = 4.37PGLDD561 pKa = 3.54TNISICQYY569 pKa = 9.87EE570 pKa = 4.2EE571 pKa = 3.67AFRR574 pKa = 11.84LVDD577 pKa = 3.79YY578 pKa = 11.6LEE580 pKa = 5.06GNPDD584 pKa = 3.56SNGYY588 pKa = 8.71WEE590 pKa = 4.68PSLPNGIFDD599 pKa = 3.86PQTNEE604 pKa = 3.56EE605 pKa = 4.1GTYY608 pKa = 9.52TYY610 pKa = 10.32HH611 pKa = 7.49ISNEE615 pKa = 3.89NCEE618 pKa = 4.25VLTSEE623 pKa = 5.42IIISFLDD630 pKa = 3.32RR631 pKa = 11.84TEE633 pKa = 4.42TISYY637 pKa = 9.36EE638 pKa = 3.93YY639 pKa = 10.04STALNNDD646 pKa = 2.42GSYY649 pKa = 11.31NIFFEE654 pKa = 4.49TANEE658 pKa = 4.01SGYY661 pKa = 9.03TFSLDD666 pKa = 4.89DD667 pKa = 4.18MTDD670 pKa = 2.88QSNGYY675 pKa = 9.21FYY677 pKa = 10.81NISPGIHH684 pKa = 5.65QIKK687 pKa = 8.0ITEE690 pKa = 4.08INGCGIVNEE699 pKa = 4.24QIALIGFPKK708 pKa = 10.31YY709 pKa = 10.6FSPNSDD715 pKa = 2.71GMNDD719 pKa = 2.9FWQPLGIIDD728 pKa = 5.29NEE730 pKa = 4.35VQTYY734 pKa = 9.97IYY736 pKa = 10.65DD737 pKa = 3.4RR738 pKa = 11.84YY739 pKa = 9.44GTLLRR744 pKa = 11.84KK745 pKa = 9.57LQGDD749 pKa = 3.93EE750 pKa = 4.41HH751 pKa = 6.95WDD753 pKa = 3.13GSYY756 pKa = 10.63NGKK759 pKa = 9.42PMPEE763 pKa = 3.5DD764 pKa = 4.69DD765 pKa = 4.11YY766 pKa = 11.47WFKK769 pKa = 11.54ALIGQEE775 pKa = 4.32SEE777 pKa = 4.4QKK779 pKa = 10.8GHH781 pKa = 6.76FSLIRR786 pKa = 4.08
Molecular weight: 87.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y1T3M7|A0A1Y1T3M7_9FLAO Uncharacterized protein OS=Zunongwangia atlantica 22II14-10F7 OX=1185767 GN=IIF7_12102 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.05RR21 pKa = 11.84MASVNGRR28 pKa = 11.84KK29 pKa = 9.21VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.73LSVSSEE47 pKa = 3.76NRR49 pKa = 11.84HH50 pKa = 4.47KK51 pKa = 10.5HH52 pKa = 4.49
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.05RR21 pKa = 11.84MASVNGRR28 pKa = 11.84KK29 pKa = 9.21VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.73LSVSSEE47 pKa = 3.76NRR49 pKa = 11.84HH50 pKa = 4.47KK51 pKa = 10.5HH52 pKa = 4.49
Molecular weight: 6.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1425727 |
29 |
2394 |
347.7 |
39.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.407 ± 0.039 | 0.649 ± 0.011 |
5.752 ± 0.031 | 7.393 ± 0.037 |
5.249 ± 0.028 | 6.377 ± 0.036 |
1.72 ± 0.02 | 7.716 ± 0.036 |
7.595 ± 0.054 | 9.194 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.182 ± 0.017 | 6.081 ± 0.041 |
3.453 ± 0.017 | 3.564 ± 0.022 |
3.625 ± 0.021 | 6.6 ± 0.034 |
5.262 ± 0.028 | 5.816 ± 0.031 |
1.147 ± 0.013 | 4.219 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
