
Sewage-associated circular DNA virus-33
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.82
Get precalculated fractions of proteins
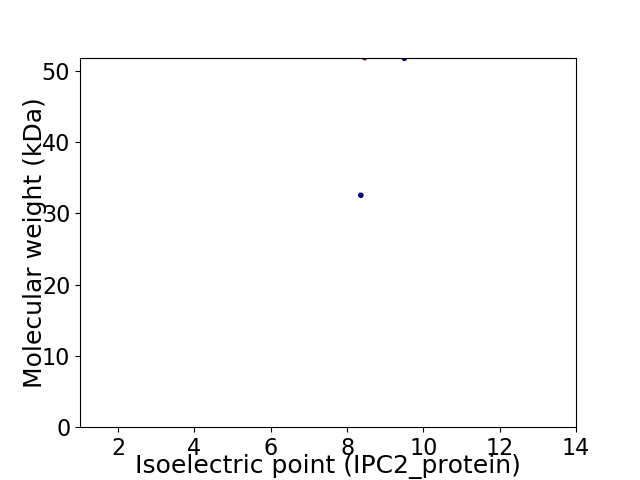
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UGP2|A0A0B4UGP2_9VIRU Capsid protein OS=Sewage-associated circular DNA virus-33 OX=1592100 PE=4 SV=1
MM1 pKa = 7.72PSSNSIQARR10 pKa = 11.84YY11 pKa = 8.33WLLTINDD18 pKa = 3.68TDD20 pKa = 5.58DD21 pKa = 3.62NVSWSPPTAINTGPWTIVQWLRR43 pKa = 11.84GQKK46 pKa = 10.26EE47 pKa = 3.63IGTNTNRR54 pKa = 11.84EE55 pKa = 3.64HH56 pKa = 5.85WQLFVAFKK64 pKa = 10.74KK65 pKa = 10.33KK66 pKa = 9.89IRR68 pKa = 11.84LSPVKK73 pKa = 10.31QLFGGARR80 pKa = 11.84VHH82 pKa = 6.83AEE84 pKa = 3.86PSRR87 pKa = 11.84SEE89 pKa = 3.73AAEE92 pKa = 3.84EE93 pKa = 4.38YY94 pKa = 10.77VFKK97 pKa = 10.81EE98 pKa = 3.97DD99 pKa = 3.1TAIANTRR106 pKa = 11.84FEE108 pKa = 4.79LGAKK112 pKa = 9.62AFNRR116 pKa = 11.84ASKK119 pKa = 9.72TDD121 pKa = 2.96WALAKK126 pKa = 10.31KK127 pKa = 9.55CAKK130 pKa = 9.54EE131 pKa = 3.93GNIDD135 pKa = 3.75GVPDD139 pKa = 3.64DD140 pKa = 5.46VYY142 pKa = 10.99IKK144 pKa = 10.76YY145 pKa = 10.9YY146 pKa = 9.57NTLKK150 pKa = 10.25TIAKK154 pKa = 10.26DD155 pKa = 3.58NMKK158 pKa = 9.92PAEE161 pKa = 4.41NLDD164 pKa = 3.83AVCGIWYY171 pKa = 8.71YY172 pKa = 11.25GPPGVGKK179 pKa = 7.53SHH181 pKa = 7.43RR182 pKa = 11.84ARR184 pKa = 11.84SEE186 pKa = 3.86FPGAYY191 pKa = 9.17MKK193 pKa = 9.38MQNKK197 pKa = 7.48WWCGYY202 pKa = 7.87QNEE205 pKa = 4.9DD206 pKa = 4.76FVILDD211 pKa = 4.49DD212 pKa = 5.14FDD214 pKa = 4.91SKK216 pKa = 11.49QLGHH220 pKa = 6.75HH221 pKa = 6.18LKK223 pKa = 9.88IWADD227 pKa = 3.17KK228 pKa = 10.16YY229 pKa = 11.67AFIAEE234 pKa = 4.4TKK236 pKa = 10.17GYY238 pKa = 10.25AINIRR243 pKa = 11.84PKK245 pKa = 10.77KK246 pKa = 10.4FIITSNYY253 pKa = 10.2SIDD256 pKa = 3.95QIFCEE261 pKa = 4.93DD262 pKa = 3.47SVLANAIKK270 pKa = 10.33RR271 pKa = 11.84RR272 pKa = 11.84FEE274 pKa = 3.74VRR276 pKa = 11.84EE277 pKa = 3.57IPLRR281 pKa = 11.84LFF283 pKa = 3.66
MM1 pKa = 7.72PSSNSIQARR10 pKa = 11.84YY11 pKa = 8.33WLLTINDD18 pKa = 3.68TDD20 pKa = 5.58DD21 pKa = 3.62NVSWSPPTAINTGPWTIVQWLRR43 pKa = 11.84GQKK46 pKa = 10.26EE47 pKa = 3.63IGTNTNRR54 pKa = 11.84EE55 pKa = 3.64HH56 pKa = 5.85WQLFVAFKK64 pKa = 10.74KK65 pKa = 10.33KK66 pKa = 9.89IRR68 pKa = 11.84LSPVKK73 pKa = 10.31QLFGGARR80 pKa = 11.84VHH82 pKa = 6.83AEE84 pKa = 3.86PSRR87 pKa = 11.84SEE89 pKa = 3.73AAEE92 pKa = 3.84EE93 pKa = 4.38YY94 pKa = 10.77VFKK97 pKa = 10.81EE98 pKa = 3.97DD99 pKa = 3.1TAIANTRR106 pKa = 11.84FEE108 pKa = 4.79LGAKK112 pKa = 9.62AFNRR116 pKa = 11.84ASKK119 pKa = 9.72TDD121 pKa = 2.96WALAKK126 pKa = 10.31KK127 pKa = 9.55CAKK130 pKa = 9.54EE131 pKa = 3.93GNIDD135 pKa = 3.75GVPDD139 pKa = 3.64DD140 pKa = 5.46VYY142 pKa = 10.99IKK144 pKa = 10.76YY145 pKa = 10.9YY146 pKa = 9.57NTLKK150 pKa = 10.25TIAKK154 pKa = 10.26DD155 pKa = 3.58NMKK158 pKa = 9.92PAEE161 pKa = 4.41NLDD164 pKa = 3.83AVCGIWYY171 pKa = 8.71YY172 pKa = 11.25GPPGVGKK179 pKa = 7.53SHH181 pKa = 7.43RR182 pKa = 11.84ARR184 pKa = 11.84SEE186 pKa = 3.86FPGAYY191 pKa = 9.17MKK193 pKa = 9.38MQNKK197 pKa = 7.48WWCGYY202 pKa = 7.87QNEE205 pKa = 4.9DD206 pKa = 4.76FVILDD211 pKa = 4.49DD212 pKa = 5.14FDD214 pKa = 4.91SKK216 pKa = 11.49QLGHH220 pKa = 6.75HH221 pKa = 6.18LKK223 pKa = 9.88IWADD227 pKa = 3.17KK228 pKa = 10.16YY229 pKa = 11.67AFIAEE234 pKa = 4.4TKK236 pKa = 10.17GYY238 pKa = 10.25AINIRR243 pKa = 11.84PKK245 pKa = 10.77KK246 pKa = 10.4FIITSNYY253 pKa = 10.2SIDD256 pKa = 3.95QIFCEE261 pKa = 4.93DD262 pKa = 3.47SVLANAIKK270 pKa = 10.33RR271 pKa = 11.84RR272 pKa = 11.84FEE274 pKa = 3.74VRR276 pKa = 11.84EE277 pKa = 3.57IPLRR281 pKa = 11.84LFF283 pKa = 3.66
Molecular weight: 32.52 kDa
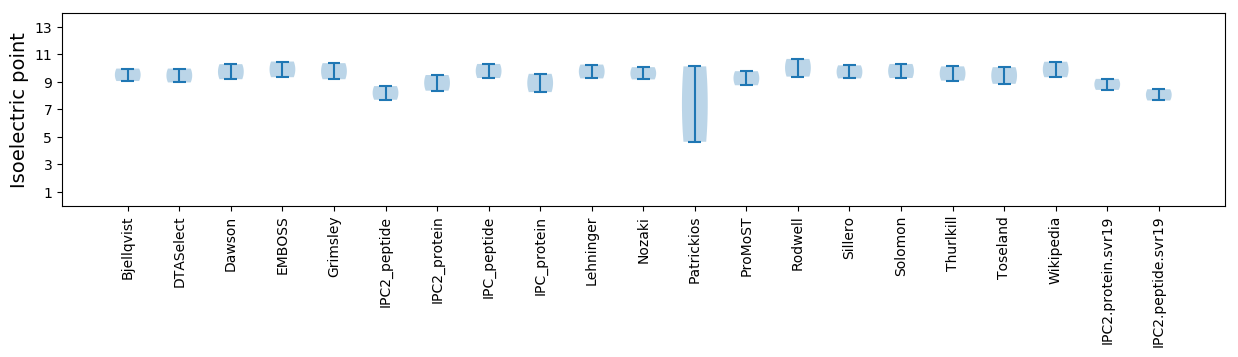
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UGP2|A0A0B4UGP2_9VIRU Capsid protein OS=Sewage-associated circular DNA virus-33 OX=1592100 PE=4 SV=1
MM1 pKa = 7.74ANRR4 pKa = 11.84AKK6 pKa = 10.29SVPRR10 pKa = 11.84TPMSLQRR17 pKa = 11.84GRR19 pKa = 11.84SRR21 pKa = 11.84SRR23 pKa = 11.84SMRR26 pKa = 11.84TVSMRR31 pKa = 11.84SATRR35 pKa = 11.84SASRR39 pKa = 11.84RR40 pKa = 11.84VLSRR44 pKa = 11.84IGSSVLGRR52 pKa = 11.84VNPYY56 pKa = 10.42VGTALSLAPYY66 pKa = 9.85ARR68 pKa = 11.84AGLRR72 pKa = 11.84TLRR75 pKa = 11.84SIRR78 pKa = 11.84KK79 pKa = 8.87YY80 pKa = 10.52SKK82 pKa = 10.37KK83 pKa = 9.89NYY85 pKa = 9.13KK86 pKa = 9.88RR87 pKa = 11.84GRR89 pKa = 11.84KK90 pKa = 8.98SGGRR94 pKa = 11.84KK95 pKa = 8.63GAAFSKK101 pKa = 10.61SAGFFGGTLKK111 pKa = 10.76DD112 pKa = 3.74SKK114 pKa = 10.58LAPYY118 pKa = 9.99LSKK121 pKa = 11.11GVVQQYY127 pKa = 10.56EE128 pKa = 4.11LGDD131 pKa = 3.87VVSEE135 pKa = 4.33ASRR138 pKa = 11.84QVVVVGHH145 pKa = 5.31STCPPSRR152 pKa = 11.84IIYY155 pKa = 10.05ACFGSLLKK163 pKa = 11.04LLFRR167 pKa = 11.84KK168 pKa = 10.19AGIKK172 pKa = 9.12IKK174 pKa = 10.41NWEE177 pKa = 3.99EE178 pKa = 4.2PILEE182 pKa = 4.43GANIPARR189 pKa = 11.84IAIRR193 pKa = 11.84YY194 pKa = 8.32KK195 pKa = 10.39EE196 pKa = 3.82RR197 pKa = 11.84DD198 pKa = 3.66GYY200 pKa = 11.51VVTTHH205 pKa = 6.69EE206 pKa = 4.42FPITTSLTMGQLVLNMVLWINGFSAINFPGQFLSIQYY243 pKa = 8.74YY244 pKa = 10.34HH245 pKa = 7.55DD246 pKa = 3.97VGTLGSSRR254 pKa = 11.84LIAYY258 pKa = 9.08DD259 pKa = 3.13IDD261 pKa = 3.92MTSTTVQIYY270 pKa = 10.02CRR272 pKa = 11.84SNMKK276 pKa = 9.52IQNRR280 pKa = 11.84TINTSGNDD288 pKa = 3.14QDD290 pKa = 5.11SDD292 pKa = 3.67VDD294 pKa = 3.89NVPLYY299 pKa = 10.8GKK301 pKa = 9.92HH302 pKa = 5.21FTVKK306 pKa = 10.77YY307 pKa = 9.67NGTVYY312 pKa = 10.6RR313 pKa = 11.84DD314 pKa = 3.64YY315 pKa = 10.68NTPAVSGTPQLYY327 pKa = 9.19TDD329 pKa = 4.51PEE331 pKa = 4.51FGSLNWSALPSDD343 pKa = 4.39TGTSLYY349 pKa = 10.86KK350 pKa = 10.2EE351 pKa = 4.45VPEE354 pKa = 4.1KK355 pKa = 11.05SQFIGVKK362 pKa = 9.81SAGPAHH368 pKa = 7.31LDD370 pKa = 3.23PGEE373 pKa = 4.23VKK375 pKa = 9.7TSKK378 pKa = 11.21LEE380 pKa = 4.03FSKK383 pKa = 11.11SLNFNKK389 pKa = 10.38LMMLFKK395 pKa = 10.9AKK397 pKa = 10.59APGGGGFPRR406 pKa = 11.84GVPFYY411 pKa = 10.67FGEE414 pKa = 4.13TRR416 pKa = 11.84FFCFEE421 pKa = 3.75KK422 pKa = 10.15MINAVAMTAEE432 pKa = 4.34NAFKK436 pKa = 10.59LAFEE440 pKa = 4.85IDD442 pKa = 3.77LTVGAICSTYY452 pKa = 10.68EE453 pKa = 3.62NHH455 pKa = 5.33QTAKK459 pKa = 9.84LTFQQTGQLL468 pKa = 3.67
MM1 pKa = 7.74ANRR4 pKa = 11.84AKK6 pKa = 10.29SVPRR10 pKa = 11.84TPMSLQRR17 pKa = 11.84GRR19 pKa = 11.84SRR21 pKa = 11.84SRR23 pKa = 11.84SMRR26 pKa = 11.84TVSMRR31 pKa = 11.84SATRR35 pKa = 11.84SASRR39 pKa = 11.84RR40 pKa = 11.84VLSRR44 pKa = 11.84IGSSVLGRR52 pKa = 11.84VNPYY56 pKa = 10.42VGTALSLAPYY66 pKa = 9.85ARR68 pKa = 11.84AGLRR72 pKa = 11.84TLRR75 pKa = 11.84SIRR78 pKa = 11.84KK79 pKa = 8.87YY80 pKa = 10.52SKK82 pKa = 10.37KK83 pKa = 9.89NYY85 pKa = 9.13KK86 pKa = 9.88RR87 pKa = 11.84GRR89 pKa = 11.84KK90 pKa = 8.98SGGRR94 pKa = 11.84KK95 pKa = 8.63GAAFSKK101 pKa = 10.61SAGFFGGTLKK111 pKa = 10.76DD112 pKa = 3.74SKK114 pKa = 10.58LAPYY118 pKa = 9.99LSKK121 pKa = 11.11GVVQQYY127 pKa = 10.56EE128 pKa = 4.11LGDD131 pKa = 3.87VVSEE135 pKa = 4.33ASRR138 pKa = 11.84QVVVVGHH145 pKa = 5.31STCPPSRR152 pKa = 11.84IIYY155 pKa = 10.05ACFGSLLKK163 pKa = 11.04LLFRR167 pKa = 11.84KK168 pKa = 10.19AGIKK172 pKa = 9.12IKK174 pKa = 10.41NWEE177 pKa = 3.99EE178 pKa = 4.2PILEE182 pKa = 4.43GANIPARR189 pKa = 11.84IAIRR193 pKa = 11.84YY194 pKa = 8.32KK195 pKa = 10.39EE196 pKa = 3.82RR197 pKa = 11.84DD198 pKa = 3.66GYY200 pKa = 11.51VVTTHH205 pKa = 6.69EE206 pKa = 4.42FPITTSLTMGQLVLNMVLWINGFSAINFPGQFLSIQYY243 pKa = 8.74YY244 pKa = 10.34HH245 pKa = 7.55DD246 pKa = 3.97VGTLGSSRR254 pKa = 11.84LIAYY258 pKa = 9.08DD259 pKa = 3.13IDD261 pKa = 3.92MTSTTVQIYY270 pKa = 10.02CRR272 pKa = 11.84SNMKK276 pKa = 9.52IQNRR280 pKa = 11.84TINTSGNDD288 pKa = 3.14QDD290 pKa = 5.11SDD292 pKa = 3.67VDD294 pKa = 3.89NVPLYY299 pKa = 10.8GKK301 pKa = 9.92HH302 pKa = 5.21FTVKK306 pKa = 10.77YY307 pKa = 9.67NGTVYY312 pKa = 10.6RR313 pKa = 11.84DD314 pKa = 3.64YY315 pKa = 10.68NTPAVSGTPQLYY327 pKa = 9.19TDD329 pKa = 4.51PEE331 pKa = 4.51FGSLNWSALPSDD343 pKa = 4.39TGTSLYY349 pKa = 10.86KK350 pKa = 10.2EE351 pKa = 4.45VPEE354 pKa = 4.1KK355 pKa = 11.05SQFIGVKK362 pKa = 9.81SAGPAHH368 pKa = 7.31LDD370 pKa = 3.23PGEE373 pKa = 4.23VKK375 pKa = 9.7TSKK378 pKa = 11.21LEE380 pKa = 4.03FSKK383 pKa = 11.11SLNFNKK389 pKa = 10.38LMMLFKK395 pKa = 10.9AKK397 pKa = 10.59APGGGGFPRR406 pKa = 11.84GVPFYY411 pKa = 10.67FGEE414 pKa = 4.13TRR416 pKa = 11.84FFCFEE421 pKa = 3.75KK422 pKa = 10.15MINAVAMTAEE432 pKa = 4.34NAFKK436 pKa = 10.59LAFEE440 pKa = 4.85IDD442 pKa = 3.77LTVGAICSTYY452 pKa = 10.68EE453 pKa = 3.62NHH455 pKa = 5.33QTAKK459 pKa = 9.84LTFQQTGQLL468 pKa = 3.67
Molecular weight: 51.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
751 |
283 |
468 |
375.5 |
42.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.856 ± 0.797 | 1.198 ± 0.129 |
4.261 ± 1.046 | 4.394 ± 0.755 |
5.06 ± 0.068 | 7.59 ± 1.16 |
1.465 ± 0.181 | 5.992 ± 1.067 |
7.457 ± 0.825 | 7.057 ± 0.841 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.13 ± 0.43 | 5.06 ± 0.567 |
4.66 ± 0.04 | 3.329 ± 0.089 |
6.258 ± 0.574 | 7.989 ± 1.611 |
6.258 ± 0.786 | 5.859 ± 0.758 |
1.731 ± 1.08 | 4.394 ± 0.092 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
