
Wilkie partiti-like virus 1
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.38
Get precalculated fractions of proteins
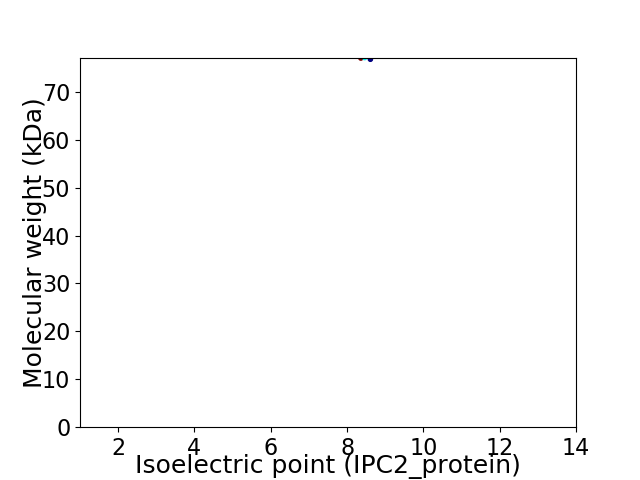
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Z2RTA3|A0A1Z2RTA3_9VIRU RdRp OS=Wilkie partiti-like virus 1 OX=2010283 PE=4 SV=1
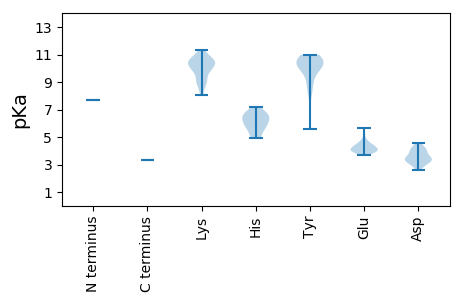
MM1 pKa = 7.71TDD3 pKa = 4.12FEE5 pKa = 4.88FFKK8 pKa = 10.55IDD10 pKa = 3.47NDD12 pKa = 4.43RR13 pKa = 11.84KK14 pKa = 10.55FEE16 pKa = 4.66DD17 pKa = 3.24NRR19 pKa = 11.84IPKK22 pKa = 9.0TGIKK26 pKa = 10.26GYY28 pKa = 8.15PQYY31 pKa = 10.98VYY33 pKa = 10.64HH34 pKa = 6.54VNQSFLQTNLEE45 pKa = 3.99QSQFGPKK52 pKa = 9.86PMTGVEE58 pKa = 4.08EE59 pKa = 4.57IIRR62 pKa = 11.84GEE64 pKa = 3.85FMEE67 pKa = 4.47YY68 pKa = 10.62TNVLYY73 pKa = 9.02TYY75 pKa = 9.09CRR77 pKa = 11.84PKK79 pKa = 10.95ADD81 pKa = 3.0VDD83 pKa = 4.17AVFNDD88 pKa = 3.87FNKK91 pKa = 8.08QTKK94 pKa = 9.01PIKK97 pKa = 10.59NLDD100 pKa = 3.6RR101 pKa = 11.84DD102 pKa = 3.76TCEE105 pKa = 3.91ATFLAIDD112 pKa = 4.23RR113 pKa = 11.84FMEE116 pKa = 3.98IQPYY120 pKa = 10.42DD121 pKa = 3.38IVHH124 pKa = 5.85WCDD127 pKa = 2.76TRR129 pKa = 11.84FYY131 pKa = 10.62KK132 pKa = 10.35WNLATKK138 pKa = 8.6VDD140 pKa = 4.57YY141 pKa = 10.9YY142 pKa = 10.84HH143 pKa = 6.62SHH145 pKa = 5.57SKK147 pKa = 8.65TRR149 pKa = 11.84LAEE152 pKa = 4.07AKK154 pKa = 10.12VNHH157 pKa = 6.67PEE159 pKa = 4.06EE160 pKa = 4.61AKK162 pKa = 10.88SPSKK166 pKa = 10.66KK167 pKa = 9.07HH168 pKa = 4.95WFINTHH174 pKa = 6.33LYY176 pKa = 10.11RR177 pKa = 11.84DD178 pKa = 3.4RR179 pKa = 11.84TVCHH183 pKa = 6.35NIKK186 pKa = 10.46LYY188 pKa = 10.61GYY190 pKa = 8.58PFVPTSGKK198 pKa = 9.36NANLEE203 pKa = 3.99LHH205 pKa = 6.16KK206 pKa = 10.38WFLKK210 pKa = 10.63HH211 pKa = 5.02PTEE214 pKa = 4.11MLVRR218 pKa = 11.84SHH220 pKa = 6.43ISKK223 pKa = 10.44RR224 pKa = 11.84EE225 pKa = 3.83KK226 pKa = 10.49LKK228 pKa = 10.73VRR230 pKa = 11.84PVYY233 pKa = 10.02CAPLLLLRR241 pKa = 11.84LEE243 pKa = 5.66CMLFWPLLAQCRR255 pKa = 11.84KK256 pKa = 9.97EE257 pKa = 4.38EE258 pKa = 3.88NCIMYY263 pKa = 10.44GLEE266 pKa = 4.22TIRR269 pKa = 11.84GGMDD273 pKa = 3.6YY274 pKa = 10.77INRR277 pKa = 11.84TATEE281 pKa = 3.92YY282 pKa = 10.64QRR284 pKa = 11.84YY285 pKa = 9.64LMLDD289 pKa = 3.37WSSFDD294 pKa = 3.12QLAPFKK300 pKa = 11.2VIDD303 pKa = 3.99LFFDD307 pKa = 3.09EE308 pKa = 4.46WLPRR312 pKa = 11.84KK313 pKa = 9.73ILVDD317 pKa = 3.32NGYY320 pKa = 10.88ALIWNYY326 pKa = 8.18QAHH329 pKa = 4.94VHH331 pKa = 6.06NFQQEE336 pKa = 4.39SKK338 pKa = 10.94AHH340 pKa = 5.57GTQSSPQPEE349 pKa = 4.25NQEE352 pKa = 3.96IPDD355 pKa = 4.1LFPNKK360 pKa = 9.19VRR362 pKa = 11.84NLISFVKK369 pKa = 9.88KK370 pKa = 9.16WYY372 pKa = 10.29KK373 pKa = 11.19DD374 pKa = 3.48MIFITPDD381 pKa = 2.59GNAYY385 pKa = 10.06RR386 pKa = 11.84RR387 pKa = 11.84INAGVPSGILMTQFLDD403 pKa = 3.09SWINLFVFSYY413 pKa = 10.56IQLKK417 pKa = 10.46SGWTMKK423 pKa = 10.06TLLANHH429 pKa = 6.92IFVMGDD435 pKa = 3.24DD436 pKa = 3.92NVIFLPTGKK445 pKa = 9.74RR446 pKa = 11.84SHH448 pKa = 5.93EE449 pKa = 3.88QLLNSITNYY458 pKa = 10.04AEE460 pKa = 4.76KK461 pKa = 10.52IFGMKK466 pKa = 10.33LNQEE470 pKa = 4.13KK471 pKa = 8.97STFTANKK478 pKa = 10.01NKK480 pKa = 10.02IEE482 pKa = 4.28VIGYY486 pKa = 5.57TNKK489 pKa = 10.41NGLPHH494 pKa = 7.21RR495 pKa = 11.84DD496 pKa = 2.91IGKK499 pKa = 9.6LVAQLAYY506 pKa = 9.69PEE508 pKa = 4.33RR509 pKa = 11.84HH510 pKa = 6.58CNDD513 pKa = 3.22EE514 pKa = 4.24DD515 pKa = 3.43MCMRR519 pKa = 11.84AVGMAYY525 pKa = 10.2ASCGVNTTFYY535 pKa = 9.0MFCKK539 pKa = 10.04RR540 pKa = 11.84VYY542 pKa = 10.11DD543 pKa = 3.83HH544 pKa = 6.8YY545 pKa = 10.96RR546 pKa = 11.84SKK548 pKa = 11.31LPLEE552 pKa = 3.89YY553 pKa = 10.4QIPFKK558 pKa = 10.4RR559 pKa = 11.84KK560 pKa = 8.25NVIGIFKK567 pKa = 10.44ALPEE571 pKa = 4.16EE572 pKa = 4.35VLSEE576 pKa = 4.19FNFVEE581 pKa = 5.24FPTLNEE587 pKa = 3.67IFCRR591 pKa = 11.84VQTHH595 pKa = 5.6QGFLDD600 pKa = 3.56IYY602 pKa = 10.09KK603 pKa = 9.99FWRR606 pKa = 11.84SDD608 pKa = 3.42YY609 pKa = 10.44FLQPPVPKK617 pKa = 9.73RR618 pKa = 11.84RR619 pKa = 11.84KK620 pKa = 8.99RR621 pKa = 11.84VTLHH625 pKa = 5.89EE626 pKa = 4.35YY627 pKa = 10.19YY628 pKa = 10.22RR629 pKa = 11.84SKK631 pKa = 10.63VWSTVTGFNRR641 pKa = 11.84QADD644 pKa = 4.15FRR646 pKa = 11.84RR647 pKa = 11.84LEE649 pKa = 4.04SS650 pKa = 3.32
MM1 pKa = 7.71TDD3 pKa = 4.12FEE5 pKa = 4.88FFKK8 pKa = 10.55IDD10 pKa = 3.47NDD12 pKa = 4.43RR13 pKa = 11.84KK14 pKa = 10.55FEE16 pKa = 4.66DD17 pKa = 3.24NRR19 pKa = 11.84IPKK22 pKa = 9.0TGIKK26 pKa = 10.26GYY28 pKa = 8.15PQYY31 pKa = 10.98VYY33 pKa = 10.64HH34 pKa = 6.54VNQSFLQTNLEE45 pKa = 3.99QSQFGPKK52 pKa = 9.86PMTGVEE58 pKa = 4.08EE59 pKa = 4.57IIRR62 pKa = 11.84GEE64 pKa = 3.85FMEE67 pKa = 4.47YY68 pKa = 10.62TNVLYY73 pKa = 9.02TYY75 pKa = 9.09CRR77 pKa = 11.84PKK79 pKa = 10.95ADD81 pKa = 3.0VDD83 pKa = 4.17AVFNDD88 pKa = 3.87FNKK91 pKa = 8.08QTKK94 pKa = 9.01PIKK97 pKa = 10.59NLDD100 pKa = 3.6RR101 pKa = 11.84DD102 pKa = 3.76TCEE105 pKa = 3.91ATFLAIDD112 pKa = 4.23RR113 pKa = 11.84FMEE116 pKa = 3.98IQPYY120 pKa = 10.42DD121 pKa = 3.38IVHH124 pKa = 5.85WCDD127 pKa = 2.76TRR129 pKa = 11.84FYY131 pKa = 10.62KK132 pKa = 10.35WNLATKK138 pKa = 8.6VDD140 pKa = 4.57YY141 pKa = 10.9YY142 pKa = 10.84HH143 pKa = 6.62SHH145 pKa = 5.57SKK147 pKa = 8.65TRR149 pKa = 11.84LAEE152 pKa = 4.07AKK154 pKa = 10.12VNHH157 pKa = 6.67PEE159 pKa = 4.06EE160 pKa = 4.61AKK162 pKa = 10.88SPSKK166 pKa = 10.66KK167 pKa = 9.07HH168 pKa = 4.95WFINTHH174 pKa = 6.33LYY176 pKa = 10.11RR177 pKa = 11.84DD178 pKa = 3.4RR179 pKa = 11.84TVCHH183 pKa = 6.35NIKK186 pKa = 10.46LYY188 pKa = 10.61GYY190 pKa = 8.58PFVPTSGKK198 pKa = 9.36NANLEE203 pKa = 3.99LHH205 pKa = 6.16KK206 pKa = 10.38WFLKK210 pKa = 10.63HH211 pKa = 5.02PTEE214 pKa = 4.11MLVRR218 pKa = 11.84SHH220 pKa = 6.43ISKK223 pKa = 10.44RR224 pKa = 11.84EE225 pKa = 3.83KK226 pKa = 10.49LKK228 pKa = 10.73VRR230 pKa = 11.84PVYY233 pKa = 10.02CAPLLLLRR241 pKa = 11.84LEE243 pKa = 5.66CMLFWPLLAQCRR255 pKa = 11.84KK256 pKa = 9.97EE257 pKa = 4.38EE258 pKa = 3.88NCIMYY263 pKa = 10.44GLEE266 pKa = 4.22TIRR269 pKa = 11.84GGMDD273 pKa = 3.6YY274 pKa = 10.77INRR277 pKa = 11.84TATEE281 pKa = 3.92YY282 pKa = 10.64QRR284 pKa = 11.84YY285 pKa = 9.64LMLDD289 pKa = 3.37WSSFDD294 pKa = 3.12QLAPFKK300 pKa = 11.2VIDD303 pKa = 3.99LFFDD307 pKa = 3.09EE308 pKa = 4.46WLPRR312 pKa = 11.84KK313 pKa = 9.73ILVDD317 pKa = 3.32NGYY320 pKa = 10.88ALIWNYY326 pKa = 8.18QAHH329 pKa = 4.94VHH331 pKa = 6.06NFQQEE336 pKa = 4.39SKK338 pKa = 10.94AHH340 pKa = 5.57GTQSSPQPEE349 pKa = 4.25NQEE352 pKa = 3.96IPDD355 pKa = 4.1LFPNKK360 pKa = 9.19VRR362 pKa = 11.84NLISFVKK369 pKa = 9.88KK370 pKa = 9.16WYY372 pKa = 10.29KK373 pKa = 11.19DD374 pKa = 3.48MIFITPDD381 pKa = 2.59GNAYY385 pKa = 10.06RR386 pKa = 11.84RR387 pKa = 11.84INAGVPSGILMTQFLDD403 pKa = 3.09SWINLFVFSYY413 pKa = 10.56IQLKK417 pKa = 10.46SGWTMKK423 pKa = 10.06TLLANHH429 pKa = 6.92IFVMGDD435 pKa = 3.24DD436 pKa = 3.92NVIFLPTGKK445 pKa = 9.74RR446 pKa = 11.84SHH448 pKa = 5.93EE449 pKa = 3.88QLLNSITNYY458 pKa = 10.04AEE460 pKa = 4.76KK461 pKa = 10.52IFGMKK466 pKa = 10.33LNQEE470 pKa = 4.13KK471 pKa = 8.97STFTANKK478 pKa = 10.01NKK480 pKa = 10.02IEE482 pKa = 4.28VIGYY486 pKa = 5.57TNKK489 pKa = 10.41NGLPHH494 pKa = 7.21RR495 pKa = 11.84DD496 pKa = 2.91IGKK499 pKa = 9.6LVAQLAYY506 pKa = 9.69PEE508 pKa = 4.33RR509 pKa = 11.84HH510 pKa = 6.58CNDD513 pKa = 3.22EE514 pKa = 4.24DD515 pKa = 3.43MCMRR519 pKa = 11.84AVGMAYY525 pKa = 10.2ASCGVNTTFYY535 pKa = 9.0MFCKK539 pKa = 10.04RR540 pKa = 11.84VYY542 pKa = 10.11DD543 pKa = 3.83HH544 pKa = 6.8YY545 pKa = 10.96RR546 pKa = 11.84SKK548 pKa = 11.31LPLEE552 pKa = 3.89YY553 pKa = 10.4QIPFKK558 pKa = 10.4RR559 pKa = 11.84KK560 pKa = 8.25NVIGIFKK567 pKa = 10.44ALPEE571 pKa = 4.16EE572 pKa = 4.35VLSEE576 pKa = 4.19FNFVEE581 pKa = 5.24FPTLNEE587 pKa = 3.67IFCRR591 pKa = 11.84VQTHH595 pKa = 5.6QGFLDD600 pKa = 3.56IYY602 pKa = 10.09KK603 pKa = 9.99FWRR606 pKa = 11.84SDD608 pKa = 3.42YY609 pKa = 10.44FLQPPVPKK617 pKa = 9.73RR618 pKa = 11.84RR619 pKa = 11.84KK620 pKa = 8.99RR621 pKa = 11.84VTLHH625 pKa = 5.89EE626 pKa = 4.35YY627 pKa = 10.19YY628 pKa = 10.22RR629 pKa = 11.84SKK631 pKa = 10.63VWSTVTGFNRR641 pKa = 11.84QADD644 pKa = 4.15FRR646 pKa = 11.84RR647 pKa = 11.84LEE649 pKa = 4.04SS650 pKa = 3.32
Molecular weight: 77.03 kDa
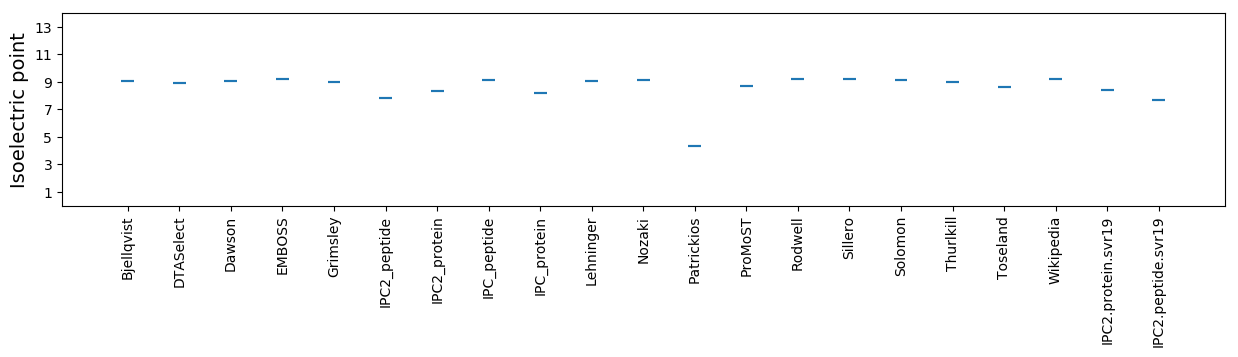
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z2RTA3|A0A1Z2RTA3_9VIRU RdRp OS=Wilkie partiti-like virus 1 OX=2010283 PE=4 SV=1
MM1 pKa = 7.71TDD3 pKa = 4.12FEE5 pKa = 4.88FFKK8 pKa = 10.55IDD10 pKa = 3.47NDD12 pKa = 4.43RR13 pKa = 11.84KK14 pKa = 10.55FEE16 pKa = 4.66DD17 pKa = 3.24NRR19 pKa = 11.84IPKK22 pKa = 9.0TGIKK26 pKa = 10.26GYY28 pKa = 8.15PQYY31 pKa = 10.98VYY33 pKa = 10.64HH34 pKa = 6.54VNQSFLQTNLEE45 pKa = 3.99QSQFGPKK52 pKa = 9.86PMTGVEE58 pKa = 4.08EE59 pKa = 4.57IIRR62 pKa = 11.84GEE64 pKa = 3.85FMEE67 pKa = 4.47YY68 pKa = 10.62TNVLYY73 pKa = 9.02TYY75 pKa = 9.09CRR77 pKa = 11.84PKK79 pKa = 10.95ADD81 pKa = 3.0VDD83 pKa = 4.17AVFNDD88 pKa = 3.87FNKK91 pKa = 8.08QTKK94 pKa = 9.01PIKK97 pKa = 10.59NLDD100 pKa = 3.6RR101 pKa = 11.84DD102 pKa = 3.76TCEE105 pKa = 3.91ATFLAIDD112 pKa = 4.23RR113 pKa = 11.84FMEE116 pKa = 3.98IQPYY120 pKa = 10.42DD121 pKa = 3.38IVHH124 pKa = 5.85WCDD127 pKa = 2.76TRR129 pKa = 11.84FYY131 pKa = 10.62KK132 pKa = 10.35WNLATKK138 pKa = 8.6VDD140 pKa = 4.57YY141 pKa = 10.9YY142 pKa = 10.84HH143 pKa = 6.62SHH145 pKa = 5.57SKK147 pKa = 8.65TRR149 pKa = 11.84LAEE152 pKa = 4.07AKK154 pKa = 10.12VNHH157 pKa = 6.67PEE159 pKa = 4.06EE160 pKa = 4.61AKK162 pKa = 10.88SPSKK166 pKa = 10.66KK167 pKa = 9.07HH168 pKa = 4.95WFINTHH174 pKa = 6.33LYY176 pKa = 10.11RR177 pKa = 11.84DD178 pKa = 3.4RR179 pKa = 11.84TVCHH183 pKa = 6.35NIKK186 pKa = 10.46LYY188 pKa = 10.61GYY190 pKa = 8.58PFVPTSGKK198 pKa = 9.36NANLEE203 pKa = 3.99LHH205 pKa = 6.16KK206 pKa = 10.38WFLKK210 pKa = 10.63HH211 pKa = 5.02PTEE214 pKa = 4.11MLVRR218 pKa = 11.84SHH220 pKa = 6.43ISKK223 pKa = 10.44RR224 pKa = 11.84EE225 pKa = 3.83KK226 pKa = 10.49LKK228 pKa = 10.73VRR230 pKa = 11.84PVYY233 pKa = 10.02CAPLLLLRR241 pKa = 11.84LEE243 pKa = 5.66CMLFWPLLAQCRR255 pKa = 11.84KK256 pKa = 9.97EE257 pKa = 4.38EE258 pKa = 3.88NCIMYY263 pKa = 10.44GLEE266 pKa = 4.22TIRR269 pKa = 11.84GGMDD273 pKa = 3.6YY274 pKa = 10.77INRR277 pKa = 11.84TATEE281 pKa = 3.92YY282 pKa = 10.64QRR284 pKa = 11.84YY285 pKa = 9.64LMLDD289 pKa = 3.37WSSFDD294 pKa = 3.12QLAPFKK300 pKa = 11.2VIDD303 pKa = 3.99LFFDD307 pKa = 3.09EE308 pKa = 4.46WLPRR312 pKa = 11.84KK313 pKa = 9.73ILVDD317 pKa = 3.32NGYY320 pKa = 10.88ALIWNYY326 pKa = 8.18QAHH329 pKa = 4.94VHH331 pKa = 6.06NFQQEE336 pKa = 4.39SKK338 pKa = 10.94AHH340 pKa = 5.57GTQSSPQPEE349 pKa = 4.25NQEE352 pKa = 3.96IPDD355 pKa = 4.1LFPNKK360 pKa = 9.19VRR362 pKa = 11.84NLISFVKK369 pKa = 9.88KK370 pKa = 9.16WYY372 pKa = 10.29KK373 pKa = 11.19DD374 pKa = 3.48MIFITPDD381 pKa = 2.59GNAYY385 pKa = 10.06RR386 pKa = 11.84RR387 pKa = 11.84INAGVPSGILMTQFLDD403 pKa = 3.09SWINLFVFSYY413 pKa = 10.56IQLKK417 pKa = 10.46SGWTMKK423 pKa = 10.06TLLANHH429 pKa = 6.92IFVMGDD435 pKa = 3.24DD436 pKa = 3.92NVIFLPTGKK445 pKa = 9.74RR446 pKa = 11.84SHH448 pKa = 5.93EE449 pKa = 3.88QLLNSITNYY458 pKa = 10.04AEE460 pKa = 4.76KK461 pKa = 10.52IFGMKK466 pKa = 10.33LNQEE470 pKa = 4.13KK471 pKa = 8.97STFTANKK478 pKa = 10.01NKK480 pKa = 10.02IEE482 pKa = 4.28VIGYY486 pKa = 5.57TNKK489 pKa = 10.41NGLPHH494 pKa = 7.21RR495 pKa = 11.84DD496 pKa = 2.91IGKK499 pKa = 9.6LVAQLAYY506 pKa = 9.69PEE508 pKa = 4.33RR509 pKa = 11.84HH510 pKa = 6.58CNDD513 pKa = 3.22EE514 pKa = 4.24DD515 pKa = 3.43MCMRR519 pKa = 11.84AVGMAYY525 pKa = 10.2ASCGVNTTFYY535 pKa = 9.0MFCKK539 pKa = 10.04RR540 pKa = 11.84VYY542 pKa = 10.11DD543 pKa = 3.83HH544 pKa = 6.8YY545 pKa = 10.96RR546 pKa = 11.84SKK548 pKa = 11.31LPLEE552 pKa = 3.89YY553 pKa = 10.4QIPFKK558 pKa = 10.4RR559 pKa = 11.84KK560 pKa = 8.25NVIGIFKK567 pKa = 10.44ALPEE571 pKa = 4.16EE572 pKa = 4.35VLSEE576 pKa = 4.19FNFVEE581 pKa = 5.24FPTLNEE587 pKa = 3.67IFCRR591 pKa = 11.84VQTHH595 pKa = 5.6QGFLDD600 pKa = 3.56IYY602 pKa = 10.09KK603 pKa = 9.99FWRR606 pKa = 11.84SDD608 pKa = 3.42YY609 pKa = 10.44FLQPPVPKK617 pKa = 9.73RR618 pKa = 11.84RR619 pKa = 11.84KK620 pKa = 8.99RR621 pKa = 11.84VTLHH625 pKa = 5.89EE626 pKa = 4.35YY627 pKa = 10.19YY628 pKa = 10.22RR629 pKa = 11.84SKK631 pKa = 10.63VWSTVTGFNRR641 pKa = 11.84QADD644 pKa = 4.15FRR646 pKa = 11.84RR647 pKa = 11.84LEE649 pKa = 4.04SS650 pKa = 3.32
MM1 pKa = 7.71TDD3 pKa = 4.12FEE5 pKa = 4.88FFKK8 pKa = 10.55IDD10 pKa = 3.47NDD12 pKa = 4.43RR13 pKa = 11.84KK14 pKa = 10.55FEE16 pKa = 4.66DD17 pKa = 3.24NRR19 pKa = 11.84IPKK22 pKa = 9.0TGIKK26 pKa = 10.26GYY28 pKa = 8.15PQYY31 pKa = 10.98VYY33 pKa = 10.64HH34 pKa = 6.54VNQSFLQTNLEE45 pKa = 3.99QSQFGPKK52 pKa = 9.86PMTGVEE58 pKa = 4.08EE59 pKa = 4.57IIRR62 pKa = 11.84GEE64 pKa = 3.85FMEE67 pKa = 4.47YY68 pKa = 10.62TNVLYY73 pKa = 9.02TYY75 pKa = 9.09CRR77 pKa = 11.84PKK79 pKa = 10.95ADD81 pKa = 3.0VDD83 pKa = 4.17AVFNDD88 pKa = 3.87FNKK91 pKa = 8.08QTKK94 pKa = 9.01PIKK97 pKa = 10.59NLDD100 pKa = 3.6RR101 pKa = 11.84DD102 pKa = 3.76TCEE105 pKa = 3.91ATFLAIDD112 pKa = 4.23RR113 pKa = 11.84FMEE116 pKa = 3.98IQPYY120 pKa = 10.42DD121 pKa = 3.38IVHH124 pKa = 5.85WCDD127 pKa = 2.76TRR129 pKa = 11.84FYY131 pKa = 10.62KK132 pKa = 10.35WNLATKK138 pKa = 8.6VDD140 pKa = 4.57YY141 pKa = 10.9YY142 pKa = 10.84HH143 pKa = 6.62SHH145 pKa = 5.57SKK147 pKa = 8.65TRR149 pKa = 11.84LAEE152 pKa = 4.07AKK154 pKa = 10.12VNHH157 pKa = 6.67PEE159 pKa = 4.06EE160 pKa = 4.61AKK162 pKa = 10.88SPSKK166 pKa = 10.66KK167 pKa = 9.07HH168 pKa = 4.95WFINTHH174 pKa = 6.33LYY176 pKa = 10.11RR177 pKa = 11.84DD178 pKa = 3.4RR179 pKa = 11.84TVCHH183 pKa = 6.35NIKK186 pKa = 10.46LYY188 pKa = 10.61GYY190 pKa = 8.58PFVPTSGKK198 pKa = 9.36NANLEE203 pKa = 3.99LHH205 pKa = 6.16KK206 pKa = 10.38WFLKK210 pKa = 10.63HH211 pKa = 5.02PTEE214 pKa = 4.11MLVRR218 pKa = 11.84SHH220 pKa = 6.43ISKK223 pKa = 10.44RR224 pKa = 11.84EE225 pKa = 3.83KK226 pKa = 10.49LKK228 pKa = 10.73VRR230 pKa = 11.84PVYY233 pKa = 10.02CAPLLLLRR241 pKa = 11.84LEE243 pKa = 5.66CMLFWPLLAQCRR255 pKa = 11.84KK256 pKa = 9.97EE257 pKa = 4.38EE258 pKa = 3.88NCIMYY263 pKa = 10.44GLEE266 pKa = 4.22TIRR269 pKa = 11.84GGMDD273 pKa = 3.6YY274 pKa = 10.77INRR277 pKa = 11.84TATEE281 pKa = 3.92YY282 pKa = 10.64QRR284 pKa = 11.84YY285 pKa = 9.64LMLDD289 pKa = 3.37WSSFDD294 pKa = 3.12QLAPFKK300 pKa = 11.2VIDD303 pKa = 3.99LFFDD307 pKa = 3.09EE308 pKa = 4.46WLPRR312 pKa = 11.84KK313 pKa = 9.73ILVDD317 pKa = 3.32NGYY320 pKa = 10.88ALIWNYY326 pKa = 8.18QAHH329 pKa = 4.94VHH331 pKa = 6.06NFQQEE336 pKa = 4.39SKK338 pKa = 10.94AHH340 pKa = 5.57GTQSSPQPEE349 pKa = 4.25NQEE352 pKa = 3.96IPDD355 pKa = 4.1LFPNKK360 pKa = 9.19VRR362 pKa = 11.84NLISFVKK369 pKa = 9.88KK370 pKa = 9.16WYY372 pKa = 10.29KK373 pKa = 11.19DD374 pKa = 3.48MIFITPDD381 pKa = 2.59GNAYY385 pKa = 10.06RR386 pKa = 11.84RR387 pKa = 11.84INAGVPSGILMTQFLDD403 pKa = 3.09SWINLFVFSYY413 pKa = 10.56IQLKK417 pKa = 10.46SGWTMKK423 pKa = 10.06TLLANHH429 pKa = 6.92IFVMGDD435 pKa = 3.24DD436 pKa = 3.92NVIFLPTGKK445 pKa = 9.74RR446 pKa = 11.84SHH448 pKa = 5.93EE449 pKa = 3.88QLLNSITNYY458 pKa = 10.04AEE460 pKa = 4.76KK461 pKa = 10.52IFGMKK466 pKa = 10.33LNQEE470 pKa = 4.13KK471 pKa = 8.97STFTANKK478 pKa = 10.01NKK480 pKa = 10.02IEE482 pKa = 4.28VIGYY486 pKa = 5.57TNKK489 pKa = 10.41NGLPHH494 pKa = 7.21RR495 pKa = 11.84DD496 pKa = 2.91IGKK499 pKa = 9.6LVAQLAYY506 pKa = 9.69PEE508 pKa = 4.33RR509 pKa = 11.84HH510 pKa = 6.58CNDD513 pKa = 3.22EE514 pKa = 4.24DD515 pKa = 3.43MCMRR519 pKa = 11.84AVGMAYY525 pKa = 10.2ASCGVNTTFYY535 pKa = 9.0MFCKK539 pKa = 10.04RR540 pKa = 11.84VYY542 pKa = 10.11DD543 pKa = 3.83HH544 pKa = 6.8YY545 pKa = 10.96RR546 pKa = 11.84SKK548 pKa = 11.31LPLEE552 pKa = 3.89YY553 pKa = 10.4QIPFKK558 pKa = 10.4RR559 pKa = 11.84KK560 pKa = 8.25NVIGIFKK567 pKa = 10.44ALPEE571 pKa = 4.16EE572 pKa = 4.35VLSEE576 pKa = 4.19FNFVEE581 pKa = 5.24FPTLNEE587 pKa = 3.67IFCRR591 pKa = 11.84VQTHH595 pKa = 5.6QGFLDD600 pKa = 3.56IYY602 pKa = 10.09KK603 pKa = 9.99FWRR606 pKa = 11.84SDD608 pKa = 3.42YY609 pKa = 10.44FLQPPVPKK617 pKa = 9.73RR618 pKa = 11.84RR619 pKa = 11.84KK620 pKa = 8.99RR621 pKa = 11.84VTLHH625 pKa = 5.89EE626 pKa = 4.35YY627 pKa = 10.19YY628 pKa = 10.22RR629 pKa = 11.84SKK631 pKa = 10.63VWSTVTGFNRR641 pKa = 11.84QADD644 pKa = 4.15FRR646 pKa = 11.84RR647 pKa = 11.84LEE649 pKa = 4.04SS650 pKa = 3.32
Molecular weight: 77.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
650 |
650 |
650 |
650.0 |
77.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.308 ± 0.0 | 2.0 ± 0.0 |
5.077 ± 0.0 | 5.846 ± 0.0 |
6.769 ± 0.0 | 4.154 ± 0.0 |
3.231 ± 0.0 | 5.846 ± 0.0 |
7.692 ± 0.0 | 8.308 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.769 ± 0.0 | 6.154 ± 0.0 |
5.077 ± 0.0 | 4.0 ± 0.0 |
5.846 ± 0.0 | 4.462 ± 0.0 |
5.538 ± 0.0 | 5.538 ± 0.0 |
2.0 ± 0.0 | 5.385 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
