
Ailuropoda melanoleuca papillomavirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Omegapapillomavirus; Omegapapillomavirus 1
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
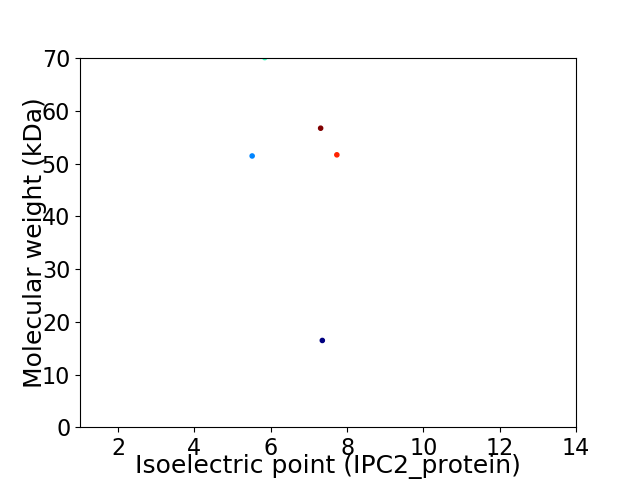
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A220IGF4|A0A220IGF4_9PAPI Minor capsid protein L2 OS=Ailuropoda melanoleuca papillomavirus 2 OX=2016455 GN=L2 PE=3 SV=1
MM1 pKa = 7.15VKK3 pKa = 10.05VLKK6 pKa = 10.18RR7 pKa = 11.84SKK9 pKa = 10.16RR10 pKa = 11.84ASAEE14 pKa = 3.65QLYY17 pKa = 7.85RR18 pKa = 11.84TCKK21 pKa = 9.55QAGTCPPDD29 pKa = 3.8VIPKK33 pKa = 9.83IEE35 pKa = 3.97QTTIADD41 pKa = 3.69QILKK45 pKa = 9.49YY46 pKa = 10.53GSSAVFFGGLGIGTGSGSGGASGYY70 pKa = 10.29VPIRR74 pKa = 11.84GGAGSSVSVGPRR86 pKa = 11.84IPIRR90 pKa = 11.84PPQIVEE96 pKa = 3.98DD97 pKa = 4.56VITIGPGDD105 pKa = 3.81SSVIGVDD112 pKa = 3.23VSPPQQPVEE121 pKa = 4.02VEE123 pKa = 4.36IIAEE127 pKa = 4.33GGGVPTVPSAADD139 pKa = 3.35TFNVVTSPDD148 pKa = 3.38TSVVSTQPPTVSTPATGASSGGRR171 pKa = 11.84AVQSSQHH178 pKa = 5.42FNNPVFEE185 pKa = 4.85ASTPRR190 pKa = 11.84APSIGEE196 pKa = 4.12TSFSPHH202 pKa = 4.35TTVVAGSSNVGVEE215 pKa = 4.55EE216 pKa = 4.12IPLAEE221 pKa = 4.78FPQSSTPLHH230 pKa = 6.15AVTRR234 pKa = 11.84GATAFTKK241 pKa = 10.23YY242 pKa = 10.43NVYY245 pKa = 9.12TRR247 pKa = 11.84QVPIEE252 pKa = 4.34SPSLYY257 pKa = 9.99TDD259 pKa = 3.37PTNLIAIRR267 pKa = 11.84NPLFDD272 pKa = 4.54IDD274 pKa = 4.89PYY276 pKa = 10.76NASATLEE283 pKa = 4.33ADD285 pKa = 3.84PDD287 pKa = 4.36NIPAPNDD294 pKa = 3.33SRR296 pKa = 11.84FLDD299 pKa = 3.28IFKK302 pKa = 10.32LHH304 pKa = 6.73RR305 pKa = 11.84PAMSRR310 pKa = 11.84TKK312 pKa = 10.29AGKK315 pKa = 9.56IRR317 pKa = 11.84FSRR320 pKa = 11.84IGFSKK325 pKa = 9.93GTVTTRR331 pKa = 11.84AGTNIGGRR339 pKa = 11.84VHH341 pKa = 7.46FYY343 pKa = 10.86HH344 pKa = 7.44DD345 pKa = 4.06FSTIVRR351 pKa = 11.84SSASEE356 pKa = 4.19SIEE359 pKa = 3.98LQPLTVGSDD368 pKa = 3.42TASSSLLSVEE378 pKa = 4.51SVGEE382 pKa = 4.3STFSSDD388 pKa = 4.61PIVSFYY394 pKa = 11.35DD395 pKa = 3.45PEE397 pKa = 6.48ADD399 pKa = 4.62LNLQLEE405 pKa = 4.69VTSDD409 pKa = 3.66SEE411 pKa = 4.22QSFVRR416 pKa = 11.84PSGGSTYY423 pKa = 10.67VDD425 pKa = 3.07NSTSVHH431 pKa = 6.09PVPNSKK437 pKa = 8.03TASHH441 pKa = 6.35TFVPSVPGFPYY452 pKa = 10.06TPIITGVFVSFDD464 pKa = 3.41FWLHH468 pKa = 6.07PSQLLKK474 pKa = 10.52RR475 pKa = 11.84KK476 pKa = 9.84RR477 pKa = 11.84SPFYY481 pKa = 10.6LADD484 pKa = 4.84GIVAAA489 pKa = 5.3
MM1 pKa = 7.15VKK3 pKa = 10.05VLKK6 pKa = 10.18RR7 pKa = 11.84SKK9 pKa = 10.16RR10 pKa = 11.84ASAEE14 pKa = 3.65QLYY17 pKa = 7.85RR18 pKa = 11.84TCKK21 pKa = 9.55QAGTCPPDD29 pKa = 3.8VIPKK33 pKa = 9.83IEE35 pKa = 3.97QTTIADD41 pKa = 3.69QILKK45 pKa = 9.49YY46 pKa = 10.53GSSAVFFGGLGIGTGSGSGGASGYY70 pKa = 10.29VPIRR74 pKa = 11.84GGAGSSVSVGPRR86 pKa = 11.84IPIRR90 pKa = 11.84PPQIVEE96 pKa = 3.98DD97 pKa = 4.56VITIGPGDD105 pKa = 3.81SSVIGVDD112 pKa = 3.23VSPPQQPVEE121 pKa = 4.02VEE123 pKa = 4.36IIAEE127 pKa = 4.33GGGVPTVPSAADD139 pKa = 3.35TFNVVTSPDD148 pKa = 3.38TSVVSTQPPTVSTPATGASSGGRR171 pKa = 11.84AVQSSQHH178 pKa = 5.42FNNPVFEE185 pKa = 4.85ASTPRR190 pKa = 11.84APSIGEE196 pKa = 4.12TSFSPHH202 pKa = 4.35TTVVAGSSNVGVEE215 pKa = 4.55EE216 pKa = 4.12IPLAEE221 pKa = 4.78FPQSSTPLHH230 pKa = 6.15AVTRR234 pKa = 11.84GATAFTKK241 pKa = 10.23YY242 pKa = 10.43NVYY245 pKa = 9.12TRR247 pKa = 11.84QVPIEE252 pKa = 4.34SPSLYY257 pKa = 9.99TDD259 pKa = 3.37PTNLIAIRR267 pKa = 11.84NPLFDD272 pKa = 4.54IDD274 pKa = 4.89PYY276 pKa = 10.76NASATLEE283 pKa = 4.33ADD285 pKa = 3.84PDD287 pKa = 4.36NIPAPNDD294 pKa = 3.33SRR296 pKa = 11.84FLDD299 pKa = 3.28IFKK302 pKa = 10.32LHH304 pKa = 6.73RR305 pKa = 11.84PAMSRR310 pKa = 11.84TKK312 pKa = 10.29AGKK315 pKa = 9.56IRR317 pKa = 11.84FSRR320 pKa = 11.84IGFSKK325 pKa = 9.93GTVTTRR331 pKa = 11.84AGTNIGGRR339 pKa = 11.84VHH341 pKa = 7.46FYY343 pKa = 10.86HH344 pKa = 7.44DD345 pKa = 4.06FSTIVRR351 pKa = 11.84SSASEE356 pKa = 4.19SIEE359 pKa = 3.98LQPLTVGSDD368 pKa = 3.42TASSSLLSVEE378 pKa = 4.51SVGEE382 pKa = 4.3STFSSDD388 pKa = 4.61PIVSFYY394 pKa = 11.35DD395 pKa = 3.45PEE397 pKa = 6.48ADD399 pKa = 4.62LNLQLEE405 pKa = 4.69VTSDD409 pKa = 3.66SEE411 pKa = 4.22QSFVRR416 pKa = 11.84PSGGSTYY423 pKa = 10.67VDD425 pKa = 3.07NSTSVHH431 pKa = 6.09PVPNSKK437 pKa = 8.03TASHH441 pKa = 6.35TFVPSVPGFPYY452 pKa = 10.06TPIITGVFVSFDD464 pKa = 3.41FWLHH468 pKa = 6.07PSQLLKK474 pKa = 10.52RR475 pKa = 11.84KK476 pKa = 9.84RR477 pKa = 11.84SPFYY481 pKa = 10.6LADD484 pKa = 4.84GIVAAA489 pKa = 5.3
Molecular weight: 51.48 kDa
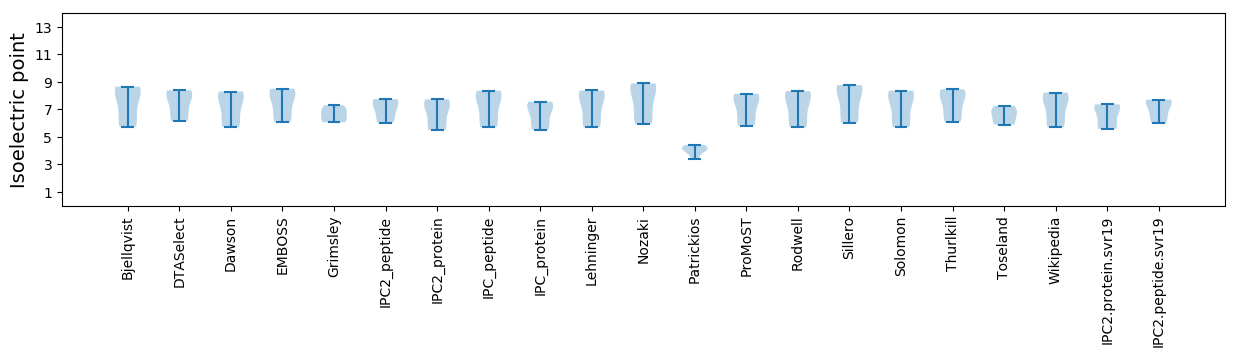
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A220IGF1|A0A220IGF1_9PAPI Replication protein E1 OS=Ailuropoda melanoleuca papillomavirus 2 OX=2016455 GN=E1 PE=3 SV=1
MM1 pKa = 7.25TKK3 pKa = 9.29MRR5 pKa = 11.84EE6 pKa = 4.06TTEE9 pKa = 3.93TLEE12 pKa = 4.06NRR14 pKa = 11.84LGALQDD20 pKa = 3.26KK21 pKa = 10.61MMDD24 pKa = 3.03IYY26 pKa = 11.27EE27 pKa = 4.47KK28 pKa = 10.72GGHH31 pKa = 5.95KK32 pKa = 10.67LEE34 pKa = 4.54DD35 pKa = 3.81QIDD38 pKa = 3.61YY39 pKa = 10.21WLLNRR44 pKa = 11.84KK45 pKa = 8.65EE46 pKa = 4.08NALLFYY52 pKa = 10.66AKK54 pKa = 10.12QKK56 pKa = 10.66GYY58 pKa = 8.45KK59 pKa = 8.78TLGLQPVPPLLVSKK73 pKa = 9.55EE74 pKa = 4.0KK75 pKa = 10.91ASIAIEE81 pKa = 3.96MHH83 pKa = 6.93LLLCSLQDD91 pKa = 3.49SEE93 pKa = 5.65YY94 pKa = 10.45GTLPWTITDD103 pKa = 3.91TSRR106 pKa = 11.84EE107 pKa = 4.07MYY109 pKa = 9.64EE110 pKa = 4.24SKK112 pKa = 10.55PEE114 pKa = 3.77KK115 pKa = 9.38TFKK118 pKa = 10.62KK119 pKa = 9.6RR120 pKa = 11.84GVHH123 pKa = 5.92VRR125 pKa = 11.84VLYY128 pKa = 10.83DD129 pKa = 4.0DD130 pKa = 4.13MQSQQVEE137 pKa = 4.45YY138 pKa = 10.5VCWLEE143 pKa = 4.55VYY145 pKa = 10.28FWDD148 pKa = 5.4CDD150 pKa = 3.56NAKK153 pKa = 9.28WGCGSSGVDD162 pKa = 2.91AHH164 pKa = 6.67GVFWTLKK171 pKa = 8.55GQKK174 pKa = 9.51KK175 pKa = 9.86YY176 pKa = 11.34YY177 pKa = 10.6EE178 pKa = 4.51NFDD181 pKa = 3.61KK182 pKa = 11.17DD183 pKa = 3.45ANKK186 pKa = 10.41YY187 pKa = 10.31GKK189 pKa = 7.96TGCWTVNFKK198 pKa = 11.3NEE200 pKa = 3.94TFRR203 pKa = 11.84FSTSASPVGKK213 pKa = 9.12PVSAAANDD221 pKa = 4.08STDD224 pKa = 2.53GRR226 pKa = 11.84APDD229 pKa = 4.11EE230 pKa = 4.62PGCPQEE236 pKa = 5.84AEE238 pKa = 4.14APVCAKK244 pKa = 10.25RR245 pKa = 11.84QADD248 pKa = 3.95GATHH252 pKa = 6.67CGEE255 pKa = 4.03APRR258 pKa = 11.84LGEE261 pKa = 3.7QQGEE265 pKa = 4.08HH266 pKa = 6.61RR267 pKa = 11.84PPEE270 pKa = 3.9KK271 pKa = 9.9RR272 pKa = 11.84RR273 pKa = 11.84RR274 pKa = 11.84ASSASEE280 pKa = 4.25GEE282 pKa = 4.07RR283 pKa = 11.84GQHH286 pKa = 4.69GKK288 pKa = 9.19QRR290 pKa = 11.84VWGRR294 pKa = 11.84RR295 pKa = 11.84QDD297 pKa = 3.73CAPSSGSVHH306 pKa = 6.11TEE308 pKa = 3.75PTCTPVQLLQQQQQQQQQRR327 pKa = 11.84QQHH330 pKa = 6.04KK331 pKa = 9.24KK332 pKa = 8.24QQLHH336 pKa = 5.29EE337 pKa = 4.21QQHH340 pKa = 4.51QQQDD344 pKa = 3.16RR345 pKa = 11.84DD346 pKa = 3.72EE347 pKa = 4.43QEE349 pKa = 3.51EE350 pKa = 4.4DD351 pKa = 3.25EE352 pKa = 5.02AGVVGDD358 pKa = 4.56PGVHH362 pKa = 6.22LSGGNTTQPASTPLPVAVLSGNANQLKK389 pKa = 9.86CYY391 pKa = 10.15RR392 pKa = 11.84YY393 pKa = 9.89RR394 pKa = 11.84LNKK397 pKa = 9.39RR398 pKa = 11.84HH399 pKa = 6.93RR400 pKa = 11.84DD401 pKa = 3.38LILYY405 pKa = 9.97ISTTWYY411 pKa = 5.4WTSNSGQHH419 pKa = 5.02KK420 pKa = 6.06TAKK423 pKa = 7.34ITVMFKK429 pKa = 9.59STACRR434 pKa = 11.84DD435 pKa = 3.5MFKK438 pKa = 10.87KK439 pKa = 10.45KK440 pKa = 10.25VVLPKK445 pKa = 10.71GVTITHH451 pKa = 6.81GAMAFF456 pKa = 3.44
MM1 pKa = 7.25TKK3 pKa = 9.29MRR5 pKa = 11.84EE6 pKa = 4.06TTEE9 pKa = 3.93TLEE12 pKa = 4.06NRR14 pKa = 11.84LGALQDD20 pKa = 3.26KK21 pKa = 10.61MMDD24 pKa = 3.03IYY26 pKa = 11.27EE27 pKa = 4.47KK28 pKa = 10.72GGHH31 pKa = 5.95KK32 pKa = 10.67LEE34 pKa = 4.54DD35 pKa = 3.81QIDD38 pKa = 3.61YY39 pKa = 10.21WLLNRR44 pKa = 11.84KK45 pKa = 8.65EE46 pKa = 4.08NALLFYY52 pKa = 10.66AKK54 pKa = 10.12QKK56 pKa = 10.66GYY58 pKa = 8.45KK59 pKa = 8.78TLGLQPVPPLLVSKK73 pKa = 9.55EE74 pKa = 4.0KK75 pKa = 10.91ASIAIEE81 pKa = 3.96MHH83 pKa = 6.93LLLCSLQDD91 pKa = 3.49SEE93 pKa = 5.65YY94 pKa = 10.45GTLPWTITDD103 pKa = 3.91TSRR106 pKa = 11.84EE107 pKa = 4.07MYY109 pKa = 9.64EE110 pKa = 4.24SKK112 pKa = 10.55PEE114 pKa = 3.77KK115 pKa = 9.38TFKK118 pKa = 10.62KK119 pKa = 9.6RR120 pKa = 11.84GVHH123 pKa = 5.92VRR125 pKa = 11.84VLYY128 pKa = 10.83DD129 pKa = 4.0DD130 pKa = 4.13MQSQQVEE137 pKa = 4.45YY138 pKa = 10.5VCWLEE143 pKa = 4.55VYY145 pKa = 10.28FWDD148 pKa = 5.4CDD150 pKa = 3.56NAKK153 pKa = 9.28WGCGSSGVDD162 pKa = 2.91AHH164 pKa = 6.67GVFWTLKK171 pKa = 8.55GQKK174 pKa = 9.51KK175 pKa = 9.86YY176 pKa = 11.34YY177 pKa = 10.6EE178 pKa = 4.51NFDD181 pKa = 3.61KK182 pKa = 11.17DD183 pKa = 3.45ANKK186 pKa = 10.41YY187 pKa = 10.31GKK189 pKa = 7.96TGCWTVNFKK198 pKa = 11.3NEE200 pKa = 3.94TFRR203 pKa = 11.84FSTSASPVGKK213 pKa = 9.12PVSAAANDD221 pKa = 4.08STDD224 pKa = 2.53GRR226 pKa = 11.84APDD229 pKa = 4.11EE230 pKa = 4.62PGCPQEE236 pKa = 5.84AEE238 pKa = 4.14APVCAKK244 pKa = 10.25RR245 pKa = 11.84QADD248 pKa = 3.95GATHH252 pKa = 6.67CGEE255 pKa = 4.03APRR258 pKa = 11.84LGEE261 pKa = 3.7QQGEE265 pKa = 4.08HH266 pKa = 6.61RR267 pKa = 11.84PPEE270 pKa = 3.9KK271 pKa = 9.9RR272 pKa = 11.84RR273 pKa = 11.84RR274 pKa = 11.84ASSASEE280 pKa = 4.25GEE282 pKa = 4.07RR283 pKa = 11.84GQHH286 pKa = 4.69GKK288 pKa = 9.19QRR290 pKa = 11.84VWGRR294 pKa = 11.84RR295 pKa = 11.84QDD297 pKa = 3.73CAPSSGSVHH306 pKa = 6.11TEE308 pKa = 3.75PTCTPVQLLQQQQQQQQQRR327 pKa = 11.84QQHH330 pKa = 6.04KK331 pKa = 9.24KK332 pKa = 8.24QQLHH336 pKa = 5.29EE337 pKa = 4.21QQHH340 pKa = 4.51QQQDD344 pKa = 3.16RR345 pKa = 11.84DD346 pKa = 3.72EE347 pKa = 4.43QEE349 pKa = 3.51EE350 pKa = 4.4DD351 pKa = 3.25EE352 pKa = 5.02AGVVGDD358 pKa = 4.56PGVHH362 pKa = 6.22LSGGNTTQPASTPLPVAVLSGNANQLKK389 pKa = 9.86CYY391 pKa = 10.15RR392 pKa = 11.84YY393 pKa = 9.89RR394 pKa = 11.84LNKK397 pKa = 9.39RR398 pKa = 11.84HH399 pKa = 6.93RR400 pKa = 11.84DD401 pKa = 3.38LILYY405 pKa = 9.97ISTTWYY411 pKa = 5.4WTSNSGQHH419 pKa = 5.02KK420 pKa = 6.06TAKK423 pKa = 7.34ITVMFKK429 pKa = 9.59STACRR434 pKa = 11.84DD435 pKa = 3.5MFKK438 pKa = 10.87KK439 pKa = 10.45KK440 pKa = 10.25VVLPKK445 pKa = 10.71GVTITHH451 pKa = 6.81GAMAFF456 pKa = 3.44
Molecular weight: 51.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2213 |
141 |
621 |
442.6 |
49.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.004 ± 0.332 | 2.44 ± 0.67 |
5.558 ± 0.262 | 5.513 ± 0.586 |
4.022 ± 0.453 | 6.914 ± 0.443 |
2.169 ± 0.267 | 3.705 ± 0.714 |
6.01 ± 0.847 | 7.727 ± 0.866 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.898 ± 0.376 | 4.338 ± 0.655 |
6.055 ± 0.901 | 4.88 ± 0.896 |
5.061 ± 0.261 | 8.134 ± 1.305 |
6.281 ± 0.698 | 7.004 ± 0.706 |
1.672 ± 0.45 | 3.615 ± 0.517 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
