
Ustilaginoidea virens RNA virus 5
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Victorivirus; unclassified Victorivirus
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
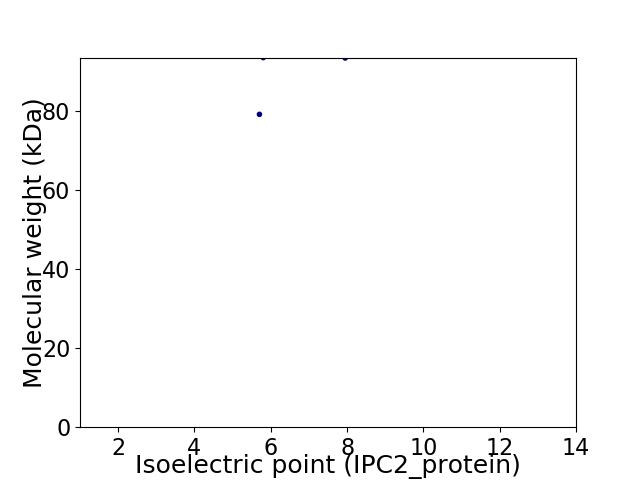
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S2UQ51|A0A0S2UQ51_9VIRU RNA-directed RNA polymerase OS=Ustilaginoidea virens RNA virus 5 OX=1756615 GN=RdRp PE=3 SV=1
MM1 pKa = 7.71ANTFNALAGVLGRR14 pKa = 11.84PRR16 pKa = 11.84TGTISGPAKK25 pKa = 8.38YY26 pKa = 9.52RR27 pKa = 11.84KK28 pKa = 8.91YY29 pKa = 9.72AASMTTSVQIRR40 pKa = 11.84GVDD43 pKa = 3.5DD44 pKa = 5.02LSVKK48 pKa = 10.4RR49 pKa = 11.84ILYY52 pKa = 9.37EE53 pKa = 3.35VGLRR57 pKa = 11.84HH58 pKa = 6.98KK59 pKa = 10.95SRR61 pKa = 11.84DD62 pKa = 3.37VALAATPEE70 pKa = 4.36TVTMIEE76 pKa = 4.0SAYY79 pKa = 10.45DD80 pKa = 3.48SSGLVSDD87 pKa = 4.44VLSGLARR94 pKa = 11.84KK95 pKa = 9.36FSNFSGTFDD104 pKa = 3.4VSNLAGVVEE113 pKa = 4.7RR114 pKa = 11.84LAKK117 pKa = 10.47GLAADD122 pKa = 3.57SAYY125 pKa = 10.46EE126 pKa = 4.32GGCTAQGLLGGNEE139 pKa = 3.97VRR141 pKa = 11.84VHH143 pKa = 6.91ALGTYY148 pKa = 7.04TGPVGAHH155 pKa = 6.99RR156 pKa = 11.84DD157 pKa = 3.56TVFIPRR163 pKa = 11.84LVDD166 pKa = 3.67SVLAPDD172 pKa = 3.9VFSVMVHH179 pKa = 5.81AVCGEE184 pKa = 4.01GGIVATDD191 pKa = 3.81LVEE194 pKa = 5.32LDD196 pKa = 3.44ANTRR200 pKa = 11.84EE201 pKa = 3.98PLIRR205 pKa = 11.84DD206 pKa = 3.21VDD208 pKa = 4.11DD209 pKa = 4.52GAFSTAVVDD218 pKa = 4.37ALRR221 pKa = 11.84LLGANFAASGQGEE234 pKa = 4.92LFSLAVVRR242 pKa = 11.84GLNAVLTVVGHH253 pKa = 5.2TDD255 pKa = 2.76EE256 pKa = 5.29GGVVRR261 pKa = 11.84DD262 pKa = 3.91VLRR265 pKa = 11.84KK266 pKa = 9.36GAFSTPYY273 pKa = 10.42GAVHH277 pKa = 7.04CGLQDD282 pKa = 3.82YY283 pKa = 9.84PGLPALASSSTSTIAGYY300 pKa = 10.42CDD302 pKa = 3.72SLLLSAAALVAHH314 pKa = 7.24CDD316 pKa = 3.23PGVVYY321 pKa = 10.8NGSWYY326 pKa = 8.02PTVLQGTSPVDD337 pKa = 3.28RR338 pKa = 11.84EE339 pKa = 4.16VRR341 pKa = 11.84PGAEE345 pKa = 4.09TPGTAEE351 pKa = 3.6MARR354 pKa = 11.84RR355 pKa = 11.84NKK357 pKa = 9.98RR358 pKa = 11.84ALLGRR363 pKa = 11.84FDD365 pKa = 3.74VFSRR369 pKa = 11.84QYY371 pKa = 9.25TAALAKK377 pKa = 10.73LFGLEE382 pKa = 4.1AGGTAASRR390 pKa = 11.84HH391 pKa = 4.61LTASASLLDD400 pKa = 4.57DD401 pKa = 4.89DD402 pKa = 5.42NRR404 pKa = 11.84HH405 pKa = 5.87LKK407 pKa = 9.42FASVSPYY414 pKa = 9.84FWVEE418 pKa = 3.52PTSIIAHH425 pKa = 6.15NFTGYY430 pKa = 9.62VAEE433 pKa = 4.58AEE435 pKa = 4.66GFASLCGRR443 pKa = 11.84GEE445 pKa = 4.29RR446 pKa = 11.84VTRR449 pKa = 11.84PAWEE453 pKa = 4.26QIAPAPGGDD462 pKa = 3.62VASSSYY468 pKa = 10.54YY469 pKa = 9.69IRR471 pKa = 11.84FRR473 pKa = 11.84GARR476 pKa = 11.84ACGYY480 pKa = 8.48LHH482 pKa = 7.0HH483 pKa = 6.62FHH485 pKa = 6.61GHH487 pKa = 6.35ANDD490 pKa = 3.21GMAFVVPRR498 pKa = 11.84QLDD501 pKa = 3.61PAGVIHH507 pKa = 6.96PGPDD511 pKa = 3.05QEE513 pKa = 4.57EE514 pKa = 4.31EE515 pKa = 3.95VRR517 pKa = 11.84DD518 pKa = 3.85RR519 pKa = 11.84LEE521 pKa = 4.16KK522 pKa = 9.85GAHH525 pKa = 5.65IGRR528 pKa = 11.84YY529 pKa = 6.79LWRR532 pKa = 11.84RR533 pKa = 11.84GQSPICAPGEE543 pKa = 3.81FLNLGEE549 pKa = 4.17TMALRR554 pKa = 11.84FRR556 pKa = 11.84HH557 pKa = 5.93LLHH560 pKa = 7.5DD561 pKa = 3.94EE562 pKa = 4.41DD563 pKa = 5.89GYY565 pKa = 11.86AVAVHH570 pKa = 6.28CPGHH574 pKa = 6.42GEE576 pKa = 4.09VEE578 pKa = 4.11ILEE581 pKa = 4.18VSYY584 pKa = 10.8AASAPIGISEE594 pKa = 4.75GPLTSAPAEE603 pKa = 3.89ARR605 pKa = 11.84RR606 pKa = 11.84ARR608 pKa = 11.84TAATRR613 pKa = 11.84ALVSACSQAQAYY625 pKa = 8.77SQATVDD631 pKa = 4.91DD632 pKa = 4.73LPLLASAPPVLRR644 pKa = 11.84RR645 pKa = 11.84AAQTPAKK652 pKa = 9.23PSEE655 pKa = 4.06GVILPGGGGKK665 pKa = 10.05LVDD668 pKa = 4.09VPSAPDD674 pKa = 3.6DD675 pKa = 3.85NEE677 pKa = 4.29AGYY680 pKa = 8.32TRR682 pKa = 11.84GTAQRR687 pKa = 11.84VVLHH691 pKa = 6.8ADD693 pKa = 3.49AVRR696 pKa = 11.84GPAVEE701 pKa = 3.95RR702 pKa = 11.84GQRR705 pKa = 11.84QGGGGVGPDD714 pKa = 3.58TPATVPRR721 pKa = 11.84PTRR724 pKa = 11.84VVVDD728 pKa = 4.18DD729 pKa = 5.21GEE731 pKa = 4.47DD732 pKa = 3.03AGAPGRR738 pKa = 11.84PALTAEE744 pKa = 4.49GATGRR749 pKa = 11.84RR750 pKa = 11.84EE751 pKa = 3.98PGRR754 pKa = 11.84AEE756 pKa = 3.74
MM1 pKa = 7.71ANTFNALAGVLGRR14 pKa = 11.84PRR16 pKa = 11.84TGTISGPAKK25 pKa = 8.38YY26 pKa = 9.52RR27 pKa = 11.84KK28 pKa = 8.91YY29 pKa = 9.72AASMTTSVQIRR40 pKa = 11.84GVDD43 pKa = 3.5DD44 pKa = 5.02LSVKK48 pKa = 10.4RR49 pKa = 11.84ILYY52 pKa = 9.37EE53 pKa = 3.35VGLRR57 pKa = 11.84HH58 pKa = 6.98KK59 pKa = 10.95SRR61 pKa = 11.84DD62 pKa = 3.37VALAATPEE70 pKa = 4.36TVTMIEE76 pKa = 4.0SAYY79 pKa = 10.45DD80 pKa = 3.48SSGLVSDD87 pKa = 4.44VLSGLARR94 pKa = 11.84KK95 pKa = 9.36FSNFSGTFDD104 pKa = 3.4VSNLAGVVEE113 pKa = 4.7RR114 pKa = 11.84LAKK117 pKa = 10.47GLAADD122 pKa = 3.57SAYY125 pKa = 10.46EE126 pKa = 4.32GGCTAQGLLGGNEE139 pKa = 3.97VRR141 pKa = 11.84VHH143 pKa = 6.91ALGTYY148 pKa = 7.04TGPVGAHH155 pKa = 6.99RR156 pKa = 11.84DD157 pKa = 3.56TVFIPRR163 pKa = 11.84LVDD166 pKa = 3.67SVLAPDD172 pKa = 3.9VFSVMVHH179 pKa = 5.81AVCGEE184 pKa = 4.01GGIVATDD191 pKa = 3.81LVEE194 pKa = 5.32LDD196 pKa = 3.44ANTRR200 pKa = 11.84EE201 pKa = 3.98PLIRR205 pKa = 11.84DD206 pKa = 3.21VDD208 pKa = 4.11DD209 pKa = 4.52GAFSTAVVDD218 pKa = 4.37ALRR221 pKa = 11.84LLGANFAASGQGEE234 pKa = 4.92LFSLAVVRR242 pKa = 11.84GLNAVLTVVGHH253 pKa = 5.2TDD255 pKa = 2.76EE256 pKa = 5.29GGVVRR261 pKa = 11.84DD262 pKa = 3.91VLRR265 pKa = 11.84KK266 pKa = 9.36GAFSTPYY273 pKa = 10.42GAVHH277 pKa = 7.04CGLQDD282 pKa = 3.82YY283 pKa = 9.84PGLPALASSSTSTIAGYY300 pKa = 10.42CDD302 pKa = 3.72SLLLSAAALVAHH314 pKa = 7.24CDD316 pKa = 3.23PGVVYY321 pKa = 10.8NGSWYY326 pKa = 8.02PTVLQGTSPVDD337 pKa = 3.28RR338 pKa = 11.84EE339 pKa = 4.16VRR341 pKa = 11.84PGAEE345 pKa = 4.09TPGTAEE351 pKa = 3.6MARR354 pKa = 11.84RR355 pKa = 11.84NKK357 pKa = 9.98RR358 pKa = 11.84ALLGRR363 pKa = 11.84FDD365 pKa = 3.74VFSRR369 pKa = 11.84QYY371 pKa = 9.25TAALAKK377 pKa = 10.73LFGLEE382 pKa = 4.1AGGTAASRR390 pKa = 11.84HH391 pKa = 4.61LTASASLLDD400 pKa = 4.57DD401 pKa = 4.89DD402 pKa = 5.42NRR404 pKa = 11.84HH405 pKa = 5.87LKK407 pKa = 9.42FASVSPYY414 pKa = 9.84FWVEE418 pKa = 3.52PTSIIAHH425 pKa = 6.15NFTGYY430 pKa = 9.62VAEE433 pKa = 4.58AEE435 pKa = 4.66GFASLCGRR443 pKa = 11.84GEE445 pKa = 4.29RR446 pKa = 11.84VTRR449 pKa = 11.84PAWEE453 pKa = 4.26QIAPAPGGDD462 pKa = 3.62VASSSYY468 pKa = 10.54YY469 pKa = 9.69IRR471 pKa = 11.84FRR473 pKa = 11.84GARR476 pKa = 11.84ACGYY480 pKa = 8.48LHH482 pKa = 7.0HH483 pKa = 6.62FHH485 pKa = 6.61GHH487 pKa = 6.35ANDD490 pKa = 3.21GMAFVVPRR498 pKa = 11.84QLDD501 pKa = 3.61PAGVIHH507 pKa = 6.96PGPDD511 pKa = 3.05QEE513 pKa = 4.57EE514 pKa = 4.31EE515 pKa = 3.95VRR517 pKa = 11.84DD518 pKa = 3.85RR519 pKa = 11.84LEE521 pKa = 4.16KK522 pKa = 9.85GAHH525 pKa = 5.65IGRR528 pKa = 11.84YY529 pKa = 6.79LWRR532 pKa = 11.84RR533 pKa = 11.84GQSPICAPGEE543 pKa = 3.81FLNLGEE549 pKa = 4.17TMALRR554 pKa = 11.84FRR556 pKa = 11.84HH557 pKa = 5.93LLHH560 pKa = 7.5DD561 pKa = 3.94EE562 pKa = 4.41DD563 pKa = 5.89GYY565 pKa = 11.86AVAVHH570 pKa = 6.28CPGHH574 pKa = 6.42GEE576 pKa = 4.09VEE578 pKa = 4.11ILEE581 pKa = 4.18VSYY584 pKa = 10.8AASAPIGISEE594 pKa = 4.75GPLTSAPAEE603 pKa = 3.89ARR605 pKa = 11.84RR606 pKa = 11.84ARR608 pKa = 11.84TAATRR613 pKa = 11.84ALVSACSQAQAYY625 pKa = 8.77SQATVDD631 pKa = 4.91DD632 pKa = 4.73LPLLASAPPVLRR644 pKa = 11.84RR645 pKa = 11.84AAQTPAKK652 pKa = 9.23PSEE655 pKa = 4.06GVILPGGGGKK665 pKa = 10.05LVDD668 pKa = 4.09VPSAPDD674 pKa = 3.6DD675 pKa = 3.85NEE677 pKa = 4.29AGYY680 pKa = 8.32TRR682 pKa = 11.84GTAQRR687 pKa = 11.84VVLHH691 pKa = 6.8ADD693 pKa = 3.49AVRR696 pKa = 11.84GPAVEE701 pKa = 3.95RR702 pKa = 11.84GQRR705 pKa = 11.84QGGGGVGPDD714 pKa = 3.58TPATVPRR721 pKa = 11.84PTRR724 pKa = 11.84VVVDD728 pKa = 4.18DD729 pKa = 5.21GEE731 pKa = 4.47DD732 pKa = 3.03AGAPGRR738 pKa = 11.84PALTAEE744 pKa = 4.49GATGRR749 pKa = 11.84RR750 pKa = 11.84EE751 pKa = 3.98PGRR754 pKa = 11.84AEE756 pKa = 3.74
Molecular weight: 79.17 kDa
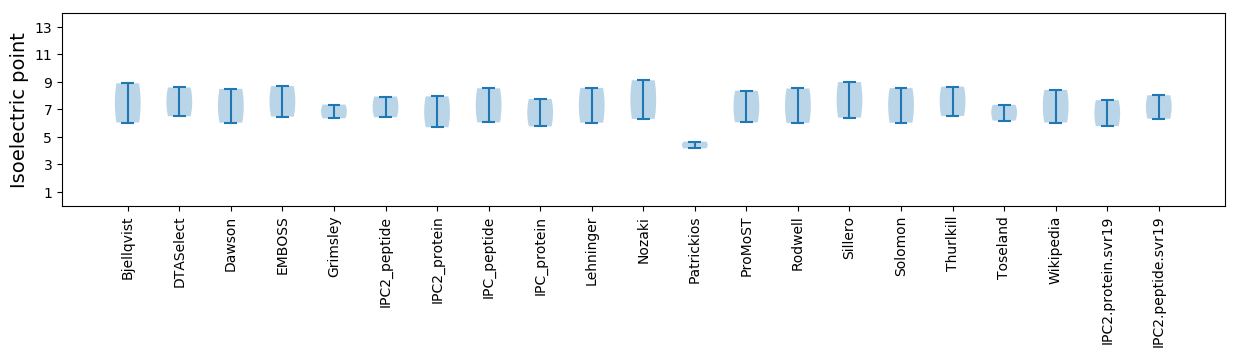
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S2UQ51|A0A0S2UQ51_9VIRU RNA-directed RNA polymerase OS=Ustilaginoidea virens RNA virus 5 OX=1756615 GN=RdRp PE=3 SV=1
MM1 pKa = 7.63AAHH4 pKa = 6.86LSISEE9 pKa = 3.79RR10 pKa = 11.84RR11 pKa = 11.84AALGHH16 pKa = 6.73LGDD19 pKa = 3.93VFYY22 pKa = 11.27GMLEE26 pKa = 3.97KK27 pKa = 11.1AEE29 pKa = 4.35FPAEE33 pKa = 3.96KK34 pKa = 9.96FRR36 pKa = 11.84EE37 pKa = 4.12LRR39 pKa = 11.84ATEE42 pKa = 3.92QLLLLSVGGGVLNSSLALAVEE63 pKa = 4.64FNRR66 pKa = 11.84RR67 pKa = 11.84VRR69 pKa = 11.84SRR71 pKa = 11.84VGLKK75 pKa = 9.51GQPGLKK81 pKa = 9.16TGVVVDD87 pKa = 3.68PLLRR91 pKa = 11.84VAASMLCARR100 pKa = 11.84FAVQVEE106 pKa = 4.45MTDD109 pKa = 3.33SNVRR113 pKa = 11.84RR114 pKa = 11.84LVRR117 pKa = 11.84LAFPEE122 pKa = 4.27ALPPLWSGVPAEE134 pKa = 4.26VRR136 pKa = 11.84GKK138 pKa = 10.83VSLSALAARR147 pKa = 11.84ADD149 pKa = 3.58LRR151 pKa = 11.84EE152 pKa = 3.67VCFPYY157 pKa = 9.29KK158 pKa = 9.85TVPQATRR165 pKa = 11.84KK166 pKa = 10.34ANVHH170 pKa = 5.96LASLLTPKK178 pKa = 10.38ARR180 pKa = 11.84EE181 pKa = 3.91LVGGLDD187 pKa = 4.79RR188 pKa = 11.84LIGWLAGRR196 pKa = 11.84CSDD199 pKa = 5.04DD200 pKa = 3.6QVCSAIIYY208 pKa = 9.29AHH210 pKa = 6.82ALGSRR215 pKa = 11.84WGPGAAEE222 pKa = 3.54IAARR226 pKa = 11.84YY227 pKa = 9.39ILDD230 pKa = 3.63PEE232 pKa = 4.64GAVSVGLVLKK242 pKa = 11.12AMGANSGPLGAALVEE257 pKa = 4.61GKK259 pKa = 9.85SLQGRR264 pKa = 11.84GVGSLDD270 pKa = 3.33LAKK273 pKa = 10.33EE274 pKa = 4.07AEE276 pKa = 4.36QRR278 pKa = 11.84CDD280 pKa = 3.85PDD282 pKa = 3.31WVAGKK287 pKa = 9.9VLHH290 pKa = 7.02CDD292 pKa = 3.5PEE294 pKa = 4.11EE295 pKa = 3.9LRR297 pKa = 11.84LVIRR301 pKa = 11.84QILSEE306 pKa = 4.05EE307 pKa = 4.04LKK309 pKa = 10.74GRR311 pKa = 11.84EE312 pKa = 4.01IVFDD316 pKa = 3.63TPEE319 pKa = 4.06QFWEE323 pKa = 5.35RR324 pKa = 11.84RR325 pKa = 11.84WQWCVNGSHH334 pKa = 6.19NRR336 pKa = 11.84TWDD339 pKa = 2.93ARR341 pKa = 11.84AGVDD345 pKa = 4.95LPASMPGCDD354 pKa = 2.73RR355 pKa = 11.84FYY357 pKa = 10.82RR358 pKa = 11.84RR359 pKa = 11.84AFSEE363 pKa = 4.3VCKK366 pKa = 11.01VEE368 pKa = 4.13TLTGWSGEE376 pKa = 4.1VLAGVSPKK384 pKa = 10.68LEE386 pKa = 4.2NGKK389 pKa = 7.79TRR391 pKa = 11.84AIFACDD397 pKa = 3.19TLSYY401 pKa = 11.01YY402 pKa = 11.0AFEE405 pKa = 4.82HH406 pKa = 6.43LLGPVSAAWLDD417 pKa = 3.56RR418 pKa = 11.84RR419 pKa = 11.84VVLDD423 pKa = 3.77PGRR426 pKa = 11.84VGHH429 pKa = 7.2LGMAEE434 pKa = 4.31RR435 pKa = 11.84INRR438 pKa = 11.84TRR440 pKa = 11.84DD441 pKa = 2.98GGGIDD446 pKa = 4.48VMLDD450 pKa = 3.21YY451 pKa = 11.55DD452 pKa = 4.93DD453 pKa = 5.9FNSHH457 pKa = 6.79HH458 pKa = 7.48SNTSMRR464 pKa = 11.84ILLEE468 pKa = 4.11EE469 pKa = 3.76TCAAVGYY476 pKa = 10.27DD477 pKa = 3.57EE478 pKa = 4.55EE479 pKa = 5.15LGRR482 pKa = 11.84KK483 pKa = 8.92LCQSFDD489 pKa = 3.74NTWVKK494 pKa = 9.83TPAGLSRR501 pKa = 11.84VRR503 pKa = 11.84GTLMSGHH510 pKa = 7.15RR511 pKa = 11.84GTTYY515 pKa = 10.57INSVLNAAYY524 pKa = 9.42IRR526 pKa = 11.84LAVGRR531 pKa = 11.84AAYY534 pKa = 9.89EE535 pKa = 4.36GMVSMHH541 pKa = 6.61VGDD544 pKa = 5.01DD545 pKa = 3.93VYY547 pKa = 11.73VNCPTPEE554 pKa = 4.08GVEE557 pKa = 4.09EE558 pKa = 4.28LVDD561 pKa = 3.46RR562 pKa = 11.84CAAIGCRR569 pKa = 11.84MNPTKK574 pKa = 10.62QSVGKK579 pKa = 10.53VGAEE583 pKa = 3.97FLRR586 pKa = 11.84MGIRR590 pKa = 11.84RR591 pKa = 11.84EE592 pKa = 4.01GAHH595 pKa = 6.75GYY597 pKa = 9.08LARR600 pKa = 11.84SVASLVSGNWVNEE613 pKa = 3.7KK614 pKa = 10.78VMDD617 pKa = 4.44PEE619 pKa = 4.36EE620 pKa = 4.65LLSSMVGTVRR630 pKa = 11.84SLINRR635 pKa = 11.84SGCEE639 pKa = 3.77TVPQLLAPAVSAVTRR654 pKa = 11.84IKK656 pKa = 10.55VARR659 pKa = 11.84CVSLLSGQSALEE671 pKa = 3.84GRR673 pKa = 11.84PVFNPRR679 pKa = 11.84DD680 pKa = 3.52GLIRR684 pKa = 11.84TWALRR689 pKa = 11.84VEE691 pKa = 4.63RR692 pKa = 11.84PASKK696 pKa = 10.64VKK698 pKa = 10.93AKK700 pKa = 10.29DD701 pKa = 3.61LPSNATDD708 pKa = 4.37DD709 pKa = 4.4YY710 pKa = 10.51LAKK713 pKa = 10.29AASVLEE719 pKa = 3.98LRR721 pKa = 11.84GAAMSTVDD729 pKa = 4.12PRR731 pKa = 11.84AAMLDD736 pKa = 3.17SSYY739 pKa = 11.31RR740 pKa = 11.84KK741 pKa = 7.6TLAGDD746 pKa = 3.7TDD748 pKa = 3.91PSKK751 pKa = 11.0IKK753 pKa = 9.77LTLRR757 pKa = 11.84SCTPVLARR765 pKa = 11.84GATNARR771 pKa = 11.84DD772 pKa = 3.92LLRR775 pKa = 11.84RR776 pKa = 11.84PCPPGALEE784 pKa = 5.36RR785 pKa = 11.84YY786 pKa = 8.62PLLQLVKK793 pKa = 10.63SGLRR797 pKa = 11.84DD798 pKa = 3.19SDD800 pKa = 3.36VRR802 pKa = 11.84EE803 pKa = 4.15LVRR806 pKa = 11.84EE807 pKa = 4.17AGGDD811 pKa = 3.04WTARR815 pKa = 11.84DD816 pKa = 3.5IGAEE820 pKa = 3.55AWGIPSRR827 pKa = 11.84SRR829 pKa = 11.84AISGVASYY837 pKa = 11.05TDD839 pKa = 3.39AASLARR845 pKa = 11.84RR846 pKa = 11.84AEE848 pKa = 3.96TDD850 pKa = 3.61VVFFPYY856 pKa = 9.13PVHH859 pKa = 5.96MM860 pKa = 5.35
MM1 pKa = 7.63AAHH4 pKa = 6.86LSISEE9 pKa = 3.79RR10 pKa = 11.84RR11 pKa = 11.84AALGHH16 pKa = 6.73LGDD19 pKa = 3.93VFYY22 pKa = 11.27GMLEE26 pKa = 3.97KK27 pKa = 11.1AEE29 pKa = 4.35FPAEE33 pKa = 3.96KK34 pKa = 9.96FRR36 pKa = 11.84EE37 pKa = 4.12LRR39 pKa = 11.84ATEE42 pKa = 3.92QLLLLSVGGGVLNSSLALAVEE63 pKa = 4.64FNRR66 pKa = 11.84RR67 pKa = 11.84VRR69 pKa = 11.84SRR71 pKa = 11.84VGLKK75 pKa = 9.51GQPGLKK81 pKa = 9.16TGVVVDD87 pKa = 3.68PLLRR91 pKa = 11.84VAASMLCARR100 pKa = 11.84FAVQVEE106 pKa = 4.45MTDD109 pKa = 3.33SNVRR113 pKa = 11.84RR114 pKa = 11.84LVRR117 pKa = 11.84LAFPEE122 pKa = 4.27ALPPLWSGVPAEE134 pKa = 4.26VRR136 pKa = 11.84GKK138 pKa = 10.83VSLSALAARR147 pKa = 11.84ADD149 pKa = 3.58LRR151 pKa = 11.84EE152 pKa = 3.67VCFPYY157 pKa = 9.29KK158 pKa = 9.85TVPQATRR165 pKa = 11.84KK166 pKa = 10.34ANVHH170 pKa = 5.96LASLLTPKK178 pKa = 10.38ARR180 pKa = 11.84EE181 pKa = 3.91LVGGLDD187 pKa = 4.79RR188 pKa = 11.84LIGWLAGRR196 pKa = 11.84CSDD199 pKa = 5.04DD200 pKa = 3.6QVCSAIIYY208 pKa = 9.29AHH210 pKa = 6.82ALGSRR215 pKa = 11.84WGPGAAEE222 pKa = 3.54IAARR226 pKa = 11.84YY227 pKa = 9.39ILDD230 pKa = 3.63PEE232 pKa = 4.64GAVSVGLVLKK242 pKa = 11.12AMGANSGPLGAALVEE257 pKa = 4.61GKK259 pKa = 9.85SLQGRR264 pKa = 11.84GVGSLDD270 pKa = 3.33LAKK273 pKa = 10.33EE274 pKa = 4.07AEE276 pKa = 4.36QRR278 pKa = 11.84CDD280 pKa = 3.85PDD282 pKa = 3.31WVAGKK287 pKa = 9.9VLHH290 pKa = 7.02CDD292 pKa = 3.5PEE294 pKa = 4.11EE295 pKa = 3.9LRR297 pKa = 11.84LVIRR301 pKa = 11.84QILSEE306 pKa = 4.05EE307 pKa = 4.04LKK309 pKa = 10.74GRR311 pKa = 11.84EE312 pKa = 4.01IVFDD316 pKa = 3.63TPEE319 pKa = 4.06QFWEE323 pKa = 5.35RR324 pKa = 11.84RR325 pKa = 11.84WQWCVNGSHH334 pKa = 6.19NRR336 pKa = 11.84TWDD339 pKa = 2.93ARR341 pKa = 11.84AGVDD345 pKa = 4.95LPASMPGCDD354 pKa = 2.73RR355 pKa = 11.84FYY357 pKa = 10.82RR358 pKa = 11.84RR359 pKa = 11.84AFSEE363 pKa = 4.3VCKK366 pKa = 11.01VEE368 pKa = 4.13TLTGWSGEE376 pKa = 4.1VLAGVSPKK384 pKa = 10.68LEE386 pKa = 4.2NGKK389 pKa = 7.79TRR391 pKa = 11.84AIFACDD397 pKa = 3.19TLSYY401 pKa = 11.01YY402 pKa = 11.0AFEE405 pKa = 4.82HH406 pKa = 6.43LLGPVSAAWLDD417 pKa = 3.56RR418 pKa = 11.84RR419 pKa = 11.84VVLDD423 pKa = 3.77PGRR426 pKa = 11.84VGHH429 pKa = 7.2LGMAEE434 pKa = 4.31RR435 pKa = 11.84INRR438 pKa = 11.84TRR440 pKa = 11.84DD441 pKa = 2.98GGGIDD446 pKa = 4.48VMLDD450 pKa = 3.21YY451 pKa = 11.55DD452 pKa = 4.93DD453 pKa = 5.9FNSHH457 pKa = 6.79HH458 pKa = 7.48SNTSMRR464 pKa = 11.84ILLEE468 pKa = 4.11EE469 pKa = 3.76TCAAVGYY476 pKa = 10.27DD477 pKa = 3.57EE478 pKa = 4.55EE479 pKa = 5.15LGRR482 pKa = 11.84KK483 pKa = 8.92LCQSFDD489 pKa = 3.74NTWVKK494 pKa = 9.83TPAGLSRR501 pKa = 11.84VRR503 pKa = 11.84GTLMSGHH510 pKa = 7.15RR511 pKa = 11.84GTTYY515 pKa = 10.57INSVLNAAYY524 pKa = 9.42IRR526 pKa = 11.84LAVGRR531 pKa = 11.84AAYY534 pKa = 9.89EE535 pKa = 4.36GMVSMHH541 pKa = 6.61VGDD544 pKa = 5.01DD545 pKa = 3.93VYY547 pKa = 11.73VNCPTPEE554 pKa = 4.08GVEE557 pKa = 4.09EE558 pKa = 4.28LVDD561 pKa = 3.46RR562 pKa = 11.84CAAIGCRR569 pKa = 11.84MNPTKK574 pKa = 10.62QSVGKK579 pKa = 10.53VGAEE583 pKa = 3.97FLRR586 pKa = 11.84MGIRR590 pKa = 11.84RR591 pKa = 11.84EE592 pKa = 4.01GAHH595 pKa = 6.75GYY597 pKa = 9.08LARR600 pKa = 11.84SVASLVSGNWVNEE613 pKa = 3.7KK614 pKa = 10.78VMDD617 pKa = 4.44PEE619 pKa = 4.36EE620 pKa = 4.65LLSSMVGTVRR630 pKa = 11.84SLINRR635 pKa = 11.84SGCEE639 pKa = 3.77TVPQLLAPAVSAVTRR654 pKa = 11.84IKK656 pKa = 10.55VARR659 pKa = 11.84CVSLLSGQSALEE671 pKa = 3.84GRR673 pKa = 11.84PVFNPRR679 pKa = 11.84DD680 pKa = 3.52GLIRR684 pKa = 11.84TWALRR689 pKa = 11.84VEE691 pKa = 4.63RR692 pKa = 11.84PASKK696 pKa = 10.64VKK698 pKa = 10.93AKK700 pKa = 10.29DD701 pKa = 3.61LPSNATDD708 pKa = 4.37DD709 pKa = 4.4YY710 pKa = 10.51LAKK713 pKa = 10.29AASVLEE719 pKa = 3.98LRR721 pKa = 11.84GAAMSTVDD729 pKa = 4.12PRR731 pKa = 11.84AAMLDD736 pKa = 3.17SSYY739 pKa = 11.31RR740 pKa = 11.84KK741 pKa = 7.6TLAGDD746 pKa = 3.7TDD748 pKa = 3.91PSKK751 pKa = 11.0IKK753 pKa = 9.77LTLRR757 pKa = 11.84SCTPVLARR765 pKa = 11.84GATNARR771 pKa = 11.84DD772 pKa = 3.92LLRR775 pKa = 11.84RR776 pKa = 11.84PCPPGALEE784 pKa = 5.36RR785 pKa = 11.84YY786 pKa = 8.62PLLQLVKK793 pKa = 10.63SGLRR797 pKa = 11.84DD798 pKa = 3.19SDD800 pKa = 3.36VRR802 pKa = 11.84EE803 pKa = 4.15LVRR806 pKa = 11.84EE807 pKa = 4.17AGGDD811 pKa = 3.04WTARR815 pKa = 11.84DD816 pKa = 3.5IGAEE820 pKa = 3.55AWGIPSRR827 pKa = 11.84SRR829 pKa = 11.84AISGVASYY837 pKa = 11.05TDD839 pKa = 3.39AASLARR845 pKa = 11.84RR846 pKa = 11.84AEE848 pKa = 3.96TDD850 pKa = 3.61VVFFPYY856 pKa = 9.13PVHH859 pKa = 5.96MM860 pKa = 5.35
Molecular weight: 93.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1616 |
756 |
860 |
808.0 |
86.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.314 ± 0.881 | 1.795 ± 0.317 |
5.446 ± 0.163 | 5.631 ± 0.318 |
2.537 ± 0.251 | 10.025 ± 0.908 |
2.166 ± 0.412 | 2.661 ± 0.099 |
2.661 ± 0.633 | 10.025 ± 0.871 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.609 ± 0.459 | 2.29 ± 0.205 |
5.446 ± 0.43 | 1.98 ± 0.181 |
8.54 ± 0.583 | 6.931 ± 0.213 |
4.95 ± 0.585 | 9.282 ± 0.074 |
1.176 ± 0.435 | 2.537 ± 0.251 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
