
Beihai sobemo-like virus 26
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
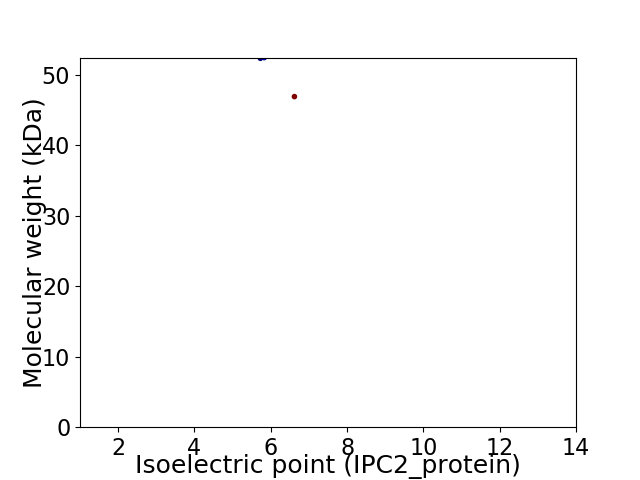
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KE73|A0A1L3KE73_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 26 OX=1922698 PE=4 SV=1
MM1 pKa = 7.83WNPDD5 pKa = 3.3LEE7 pKa = 4.39ATEE10 pKa = 4.95LVCSSWADD18 pKa = 3.27VEE20 pKa = 4.27RR21 pKa = 11.84ASFFRR26 pKa = 11.84DD27 pKa = 5.11FIQPHH32 pKa = 5.75LTQDD36 pKa = 3.42NLNSVLLVVATASVLSFLQYY56 pKa = 11.39ASILFVRR63 pKa = 11.84RR64 pKa = 11.84CRR66 pKa = 11.84SARR69 pKa = 11.84SLFFEE74 pKa = 4.61SVQAGAPFYY83 pKa = 11.02SSGEE87 pKa = 4.09SPPCQVEE94 pKa = 3.94MRR96 pKa = 11.84KK97 pKa = 10.02GSRR100 pKa = 11.84LVGFGIRR107 pKa = 11.84VGDD110 pKa = 4.13WIVTPQHH117 pKa = 6.11VADD120 pKa = 4.4SSDD123 pKa = 3.71NVWGGTSMFTFEE135 pKa = 5.37NYY137 pKa = 7.76STLCTDD143 pKa = 3.68AVAIRR148 pKa = 11.84IPANWNDD155 pKa = 3.1VRR157 pKa = 11.84KK158 pKa = 10.05ASVGALCKK166 pKa = 10.47NLIVRR171 pKa = 11.84CVGPMGEE178 pKa = 4.22WSCGRR183 pKa = 11.84LAEE186 pKa = 4.57ADD188 pKa = 3.17LWGGLDD194 pKa = 3.62YY195 pKa = 11.12FGSTDD200 pKa = 3.22HH201 pKa = 6.58GFSGAAYY208 pKa = 9.58ASGNTVYY215 pKa = 11.03GMHH218 pKa = 7.03CGGSGPNKK226 pKa = 10.18PNFGWSLSFLSCFVDD241 pKa = 5.36DD242 pKa = 4.39LAEE245 pKa = 4.08EE246 pKa = 4.15QRR248 pKa = 11.84TPVYY252 pKa = 9.9EE253 pKa = 4.13EE254 pKa = 3.77PSVRR258 pKa = 11.84EE259 pKa = 3.94VTTQAGKK266 pKa = 10.68RR267 pKa = 11.84KK268 pKa = 9.24IKK270 pKa = 10.06KK271 pKa = 8.88DD272 pKa = 2.83QGRR275 pKa = 11.84FVRR278 pKa = 11.84YY279 pKa = 6.12TQEE282 pKa = 3.82PTPVSVGRR290 pKa = 11.84HH291 pKa = 4.2VDD293 pKa = 3.2SSIVDD298 pKa = 3.66DD299 pKa = 4.23VAANADD305 pKa = 3.38NWFKK309 pKa = 11.43DD310 pKa = 3.37LGYY313 pKa = 10.41EE314 pKa = 3.95AALVEE319 pKa = 4.8SITKK323 pKa = 10.32KK324 pKa = 10.81VMNEE328 pKa = 3.53LSGNGLGPLRR338 pKa = 11.84GASTVSAVASPTSKK352 pKa = 10.5SSEE355 pKa = 3.97EE356 pKa = 4.13KK357 pKa = 10.7APSPAADD364 pKa = 3.39AQLPSTAQQSSPSPQLRR381 pKa = 11.84QTSTVTTVPEE391 pKa = 4.09KK392 pKa = 10.4LSKK395 pKa = 10.47RR396 pKa = 11.84MKK398 pKa = 10.59KK399 pKa = 9.52EE400 pKa = 3.47FQAAQVEE407 pKa = 4.35QLAWLRR413 pKa = 11.84EE414 pKa = 3.97TLTQRR419 pKa = 11.84FGFPVAALDD428 pKa = 3.7TTGWDD433 pKa = 3.53LQRR436 pKa = 11.84LQSHH440 pKa = 5.63VFVLQALEE448 pKa = 3.92AQSRR452 pKa = 11.84GKK454 pKa = 10.34VLSCTPASCPATDD467 pKa = 3.9QSQQGASKK475 pKa = 10.59SEE477 pKa = 4.03PSTGRR482 pKa = 3.16
MM1 pKa = 7.83WNPDD5 pKa = 3.3LEE7 pKa = 4.39ATEE10 pKa = 4.95LVCSSWADD18 pKa = 3.27VEE20 pKa = 4.27RR21 pKa = 11.84ASFFRR26 pKa = 11.84DD27 pKa = 5.11FIQPHH32 pKa = 5.75LTQDD36 pKa = 3.42NLNSVLLVVATASVLSFLQYY56 pKa = 11.39ASILFVRR63 pKa = 11.84RR64 pKa = 11.84CRR66 pKa = 11.84SARR69 pKa = 11.84SLFFEE74 pKa = 4.61SVQAGAPFYY83 pKa = 11.02SSGEE87 pKa = 4.09SPPCQVEE94 pKa = 3.94MRR96 pKa = 11.84KK97 pKa = 10.02GSRR100 pKa = 11.84LVGFGIRR107 pKa = 11.84VGDD110 pKa = 4.13WIVTPQHH117 pKa = 6.11VADD120 pKa = 4.4SSDD123 pKa = 3.71NVWGGTSMFTFEE135 pKa = 5.37NYY137 pKa = 7.76STLCTDD143 pKa = 3.68AVAIRR148 pKa = 11.84IPANWNDD155 pKa = 3.1VRR157 pKa = 11.84KK158 pKa = 10.05ASVGALCKK166 pKa = 10.47NLIVRR171 pKa = 11.84CVGPMGEE178 pKa = 4.22WSCGRR183 pKa = 11.84LAEE186 pKa = 4.57ADD188 pKa = 3.17LWGGLDD194 pKa = 3.62YY195 pKa = 11.12FGSTDD200 pKa = 3.22HH201 pKa = 6.58GFSGAAYY208 pKa = 9.58ASGNTVYY215 pKa = 11.03GMHH218 pKa = 7.03CGGSGPNKK226 pKa = 10.18PNFGWSLSFLSCFVDD241 pKa = 5.36DD242 pKa = 4.39LAEE245 pKa = 4.08EE246 pKa = 4.15QRR248 pKa = 11.84TPVYY252 pKa = 9.9EE253 pKa = 4.13EE254 pKa = 3.77PSVRR258 pKa = 11.84EE259 pKa = 3.94VTTQAGKK266 pKa = 10.68RR267 pKa = 11.84KK268 pKa = 9.24IKK270 pKa = 10.06KK271 pKa = 8.88DD272 pKa = 2.83QGRR275 pKa = 11.84FVRR278 pKa = 11.84YY279 pKa = 6.12TQEE282 pKa = 3.82PTPVSVGRR290 pKa = 11.84HH291 pKa = 4.2VDD293 pKa = 3.2SSIVDD298 pKa = 3.66DD299 pKa = 4.23VAANADD305 pKa = 3.38NWFKK309 pKa = 11.43DD310 pKa = 3.37LGYY313 pKa = 10.41EE314 pKa = 3.95AALVEE319 pKa = 4.8SITKK323 pKa = 10.32KK324 pKa = 10.81VMNEE328 pKa = 3.53LSGNGLGPLRR338 pKa = 11.84GASTVSAVASPTSKK352 pKa = 10.5SSEE355 pKa = 3.97EE356 pKa = 4.13KK357 pKa = 10.7APSPAADD364 pKa = 3.39AQLPSTAQQSSPSPQLRR381 pKa = 11.84QTSTVTTVPEE391 pKa = 4.09KK392 pKa = 10.4LSKK395 pKa = 10.47RR396 pKa = 11.84MKK398 pKa = 10.59KK399 pKa = 9.52EE400 pKa = 3.47FQAAQVEE407 pKa = 4.35QLAWLRR413 pKa = 11.84EE414 pKa = 3.97TLTQRR419 pKa = 11.84FGFPVAALDD428 pKa = 3.7TTGWDD433 pKa = 3.53LQRR436 pKa = 11.84LQSHH440 pKa = 5.63VFVLQALEE448 pKa = 3.92AQSRR452 pKa = 11.84GKK454 pKa = 10.34VLSCTPASCPATDD467 pKa = 3.9QSQQGASKK475 pKa = 10.59SEE477 pKa = 4.03PSTGRR482 pKa = 3.16
Molecular weight: 52.36 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KE73|A0A1L3KE73_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 26 OX=1922698 PE=4 SV=1
MM1 pKa = 7.35GLAAAAKK8 pKa = 10.04SRR10 pKa = 11.84FRR12 pKa = 11.84APGVGGSVEE21 pKa = 4.17RR22 pKa = 11.84KK23 pKa = 9.67SLEE26 pKa = 3.75LHH28 pKa = 6.55ASLMSCDD35 pKa = 3.34RR36 pKa = 11.84PVPARR41 pKa = 11.84SLQEE45 pKa = 4.0RR46 pKa = 11.84AFNWALRR53 pKa = 11.84EE54 pKa = 3.87LAPVCCEE61 pKa = 4.03LPPDD65 pKa = 3.99FFEE68 pKa = 5.21RR69 pKa = 11.84SHH71 pKa = 7.19FDD73 pKa = 3.11RR74 pKa = 11.84VLNSLNMQASPGYY87 pKa = 8.97PLMRR91 pKa = 11.84VAPTVGSYY99 pKa = 10.65LKK101 pKa = 10.64RR102 pKa = 11.84PDD104 pKa = 3.52GSWRR108 pKa = 11.84QDD110 pKa = 2.81RR111 pKa = 11.84VEE113 pKa = 3.98QLWASVLQRR122 pKa = 11.84VDD124 pKa = 3.44SDD126 pKa = 3.84PDD128 pKa = 4.14LIRR131 pKa = 11.84VFVKK135 pKa = 10.11QEE137 pKa = 3.4PHH139 pKa = 6.79RR140 pKa = 11.84IAKK143 pKa = 9.16VRR145 pKa = 11.84EE146 pKa = 3.58GRR148 pKa = 11.84MRR150 pKa = 11.84LIQAVSLVDD159 pKa = 3.37QVVDD163 pKa = 4.02HH164 pKa = 6.92LLHH167 pKa = 7.18DD168 pKa = 4.34HH169 pKa = 6.33LQQRR173 pKa = 11.84EE174 pKa = 3.85IEE176 pKa = 4.4LFGKK180 pKa = 9.62VPVMTGWAPVHH191 pKa = 5.93GGWRR195 pKa = 11.84HH196 pKa = 5.87LHH198 pKa = 6.01ADD200 pKa = 2.26WWGYY204 pKa = 10.45DD205 pKa = 3.12RR206 pKa = 11.84SAWDD210 pKa = 3.11WTVPAWLLRR219 pKa = 11.84LEE221 pKa = 3.99WRR223 pKa = 11.84LRR225 pKa = 11.84EE226 pKa = 4.08ALTINPTRR234 pKa = 11.84LWLSQMQKK242 pKa = 10.34RR243 pKa = 11.84YY244 pKa = 10.72DD245 pKa = 3.66EE246 pKa = 5.61LFVCPEE252 pKa = 4.23FVLSDD257 pKa = 3.4GMVVKK262 pKa = 10.17QCIEE266 pKa = 3.97GVMKK270 pKa = 10.2TGSVVTASANSHH282 pKa = 5.11MQLLLHH288 pKa = 6.73AVAAFAAGEE297 pKa = 4.24EE298 pKa = 4.26PGPLIAMGDD307 pKa = 3.76DD308 pKa = 3.35TMQPLGSEE316 pKa = 4.69AYY318 pKa = 9.84LQEE321 pKa = 3.42LRR323 pKa = 11.84KK324 pKa = 9.44YY325 pKa = 10.44VNVKK329 pKa = 10.02EE330 pKa = 4.29PEE332 pKa = 4.06LGEE335 pKa = 3.95FCSRR339 pKa = 11.84VIQGSRR345 pKa = 11.84VEE347 pKa = 4.03PQNILKK353 pKa = 10.26HH354 pKa = 4.73VANLITAPPEE364 pKa = 3.99FLDD367 pKa = 5.85DD368 pKa = 3.98MLLSLQVEE376 pKa = 4.7YY377 pKa = 10.99ARR379 pKa = 11.84SRR381 pKa = 11.84WLRR384 pKa = 11.84LWQLTARR391 pKa = 11.84RR392 pKa = 11.84LGCRR396 pKa = 11.84VRR398 pKa = 11.84PQIWLLDD405 pKa = 3.97VYY407 pKa = 11.46DD408 pKa = 4.03EE409 pKa = 4.37AA410 pKa = 6.63
MM1 pKa = 7.35GLAAAAKK8 pKa = 10.04SRR10 pKa = 11.84FRR12 pKa = 11.84APGVGGSVEE21 pKa = 4.17RR22 pKa = 11.84KK23 pKa = 9.67SLEE26 pKa = 3.75LHH28 pKa = 6.55ASLMSCDD35 pKa = 3.34RR36 pKa = 11.84PVPARR41 pKa = 11.84SLQEE45 pKa = 4.0RR46 pKa = 11.84AFNWALRR53 pKa = 11.84EE54 pKa = 3.87LAPVCCEE61 pKa = 4.03LPPDD65 pKa = 3.99FFEE68 pKa = 5.21RR69 pKa = 11.84SHH71 pKa = 7.19FDD73 pKa = 3.11RR74 pKa = 11.84VLNSLNMQASPGYY87 pKa = 8.97PLMRR91 pKa = 11.84VAPTVGSYY99 pKa = 10.65LKK101 pKa = 10.64RR102 pKa = 11.84PDD104 pKa = 3.52GSWRR108 pKa = 11.84QDD110 pKa = 2.81RR111 pKa = 11.84VEE113 pKa = 3.98QLWASVLQRR122 pKa = 11.84VDD124 pKa = 3.44SDD126 pKa = 3.84PDD128 pKa = 4.14LIRR131 pKa = 11.84VFVKK135 pKa = 10.11QEE137 pKa = 3.4PHH139 pKa = 6.79RR140 pKa = 11.84IAKK143 pKa = 9.16VRR145 pKa = 11.84EE146 pKa = 3.58GRR148 pKa = 11.84MRR150 pKa = 11.84LIQAVSLVDD159 pKa = 3.37QVVDD163 pKa = 4.02HH164 pKa = 6.92LLHH167 pKa = 7.18DD168 pKa = 4.34HH169 pKa = 6.33LQQRR173 pKa = 11.84EE174 pKa = 3.85IEE176 pKa = 4.4LFGKK180 pKa = 9.62VPVMTGWAPVHH191 pKa = 5.93GGWRR195 pKa = 11.84HH196 pKa = 5.87LHH198 pKa = 6.01ADD200 pKa = 2.26WWGYY204 pKa = 10.45DD205 pKa = 3.12RR206 pKa = 11.84SAWDD210 pKa = 3.11WTVPAWLLRR219 pKa = 11.84LEE221 pKa = 3.99WRR223 pKa = 11.84LRR225 pKa = 11.84EE226 pKa = 4.08ALTINPTRR234 pKa = 11.84LWLSQMQKK242 pKa = 10.34RR243 pKa = 11.84YY244 pKa = 10.72DD245 pKa = 3.66EE246 pKa = 5.61LFVCPEE252 pKa = 4.23FVLSDD257 pKa = 3.4GMVVKK262 pKa = 10.17QCIEE266 pKa = 3.97GVMKK270 pKa = 10.2TGSVVTASANSHH282 pKa = 5.11MQLLLHH288 pKa = 6.73AVAAFAAGEE297 pKa = 4.24EE298 pKa = 4.26PGPLIAMGDD307 pKa = 3.76DD308 pKa = 3.35TMQPLGSEE316 pKa = 4.69AYY318 pKa = 9.84LQEE321 pKa = 3.42LRR323 pKa = 11.84KK324 pKa = 9.44YY325 pKa = 10.44VNVKK329 pKa = 10.02EE330 pKa = 4.29PEE332 pKa = 4.06LGEE335 pKa = 3.95FCSRR339 pKa = 11.84VIQGSRR345 pKa = 11.84VEE347 pKa = 4.03PQNILKK353 pKa = 10.26HH354 pKa = 4.73VANLITAPPEE364 pKa = 3.99FLDD367 pKa = 5.85DD368 pKa = 3.98MLLSLQVEE376 pKa = 4.7YY377 pKa = 10.99ARR379 pKa = 11.84SRR381 pKa = 11.84WLRR384 pKa = 11.84LWQLTARR391 pKa = 11.84RR392 pKa = 11.84LGCRR396 pKa = 11.84VRR398 pKa = 11.84PQIWLLDD405 pKa = 3.97VYY407 pKa = 11.46DD408 pKa = 4.03EE409 pKa = 4.37AA410 pKa = 6.63
Molecular weight: 46.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
892 |
410 |
482 |
446.0 |
49.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.857 ± 0.345 | 2.018 ± 0.19 |
5.157 ± 0.128 | 5.942 ± 0.394 |
3.812 ± 0.541 | 6.614 ± 0.614 |
2.018 ± 0.556 | 2.354 ± 0.201 |
3.475 ± 0.335 | 9.865 ± 1.574 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.242 ± 0.568 | 2.578 ± 0.384 |
5.717 ± 0.232 | 5.269 ± 0.09 |
7.175 ± 1.131 | 8.857 ± 1.687 |
4.484 ± 1.251 | 8.744 ± 0.171 |
2.915 ± 0.455 | 1.906 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
