
Pantoea cypripedii (Pectobacterium cypripedii) (Erwinia cypripedii)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Erwiniaceae; Pantoea
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
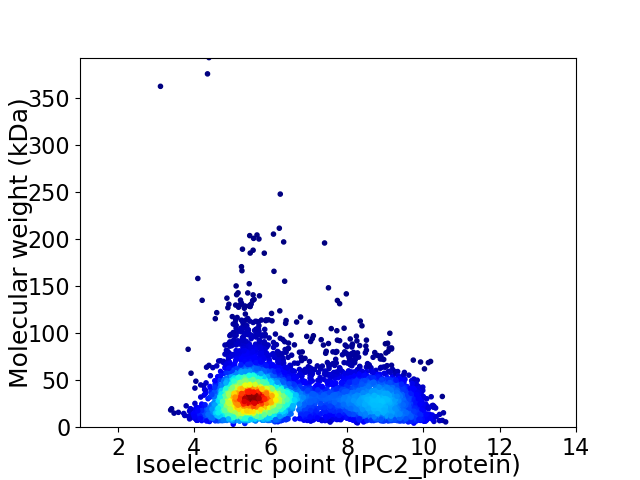
Virtual 2D-PAGE plot for 5848 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X1EY03|A0A1X1EY03_PANCY ATP-binding protein OS=Pantoea cypripedii OX=55209 GN=HA50_16835 PE=4 SV=1
MM1 pKa = 7.64LGTVTAGSNGAWSFTTSTLADD22 pKa = 3.54GTHH25 pKa = 6.16TLNVTATDD33 pKa = 3.4AAGNVSPNASVTLTVDD49 pKa = 3.21TGVPTASTLTVTADD63 pKa = 3.1NVTPNVTIPSGGYY76 pKa = 8.76TNDD79 pKa = 3.3NTPTLSGTAEE89 pKa = 4.0AGAVVTISDD98 pKa = 3.9GSTVLGSVTAGAGGAWSFTTSALSNGTHH126 pKa = 6.82PLSVTVKK133 pKa = 9.97DD134 pKa = 3.39AAGNVSPATAATVIVDD150 pKa = 4.11TVAPTASTLTITADD164 pKa = 3.12NVTPNVTVPSGGSTNDD180 pKa = 3.29TTPTLSGTAEE190 pKa = 4.0IGSKK194 pKa = 7.88VTILDD199 pKa = 3.58GSTVLGTLTVGGSGTWSFTTATLSNGTHH227 pKa = 6.75PLSVTVTDD235 pKa = 4.04LAGNVSGTTSASVIIDD251 pKa = 3.71TVAPAAVTGLAASNNNGGSAVAIPNNGTTNDD282 pKa = 3.75NTPLLSGTAEE292 pKa = 4.01AGAKK296 pKa = 9.69ISIYY300 pKa = 10.87DD301 pKa = 3.58GATLLGTTTAASSGAWSFTTSTLTEE326 pKa = 4.22GTHH329 pKa = 5.25TLNVTATDD337 pKa = 3.4AAGNVSPNASVTLTVDD353 pKa = 3.5SVAPTNSTLTVTNDD367 pKa = 2.97SVTPNVTVPSGGYY380 pKa = 8.02TRR382 pKa = 11.84DD383 pKa = 3.38TTPVLTGTAEE393 pKa = 3.79VGSRR397 pKa = 11.84VTIYY401 pKa = 10.93DD402 pKa = 3.33GATILGSVTVGAGGTWSFTTAVLANGSHH430 pKa = 6.92PLSVTVTDD438 pKa = 3.57AAGNVSGITSSTVNVDD454 pKa = 3.42TVAPTAATALAVNTAGTTLTGTGEE478 pKa = 4.3AGTTVTVKK486 pKa = 10.73DD487 pKa = 3.75AGGNTIGTGTVGVTGSFSVTLTTAQTTGATLSVSLTDD524 pKa = 3.63AAGNTSPTSTVLGAIKK540 pKa = 10.02IVAVNDD546 pKa = 3.93LNEE549 pKa = 4.61LDD551 pKa = 4.01YY552 pKa = 10.92STTSGTITNTTGTTNTGTALLSVNLGSVLGVGVLSSSNAYY592 pKa = 8.25MFNVGTGDD600 pKa = 3.28TRR602 pKa = 11.84TVTLHH607 pKa = 6.73ASEE610 pKa = 4.69TSLISLIGSYY620 pKa = 10.5SLYY623 pKa = 10.47LYY625 pKa = 9.2QQQTDD630 pKa = 4.17GTWKK634 pKa = 10.68VISTTTNYY642 pKa = 10.01ISTTLLSLLTQTGSNITYY660 pKa = 10.69SNLGTGNYY668 pKa = 10.02AVVLGASSGISVLPSTSVTTVSDD691 pKa = 3.1ITTIAATVAATVTGNLLTNDD711 pKa = 3.65TSSVAGTVPTGTAVTSVSGHH731 pKa = 4.76TVGTSTSFSTAYY743 pKa = 8.38GTLTIDD749 pKa = 2.94SHH751 pKa = 7.59GNYY754 pKa = 9.57TYY756 pKa = 10.06TLKK759 pKa = 10.98AGLDD763 pKa = 3.47AGTLPVADD771 pKa = 3.86TFSYY775 pKa = 11.02SVTDD779 pKa = 3.57AQGVVTTATLTIDD792 pKa = 4.09LVHH795 pKa = 6.33GTTTASLLSVNSLFAEE811 pKa = 4.41ATTTTDD817 pKa = 2.73SDD819 pKa = 3.95HH820 pKa = 6.77SVSGSIWADD829 pKa = 2.66SSTSHH834 pKa = 6.68TGTLTITNEE843 pKa = 4.38HH844 pKa = 6.82GDD846 pKa = 3.74STT848 pKa = 4.46
MM1 pKa = 7.64LGTVTAGSNGAWSFTTSTLADD22 pKa = 3.54GTHH25 pKa = 6.16TLNVTATDD33 pKa = 3.4AAGNVSPNASVTLTVDD49 pKa = 3.21TGVPTASTLTVTADD63 pKa = 3.1NVTPNVTIPSGGYY76 pKa = 8.76TNDD79 pKa = 3.3NTPTLSGTAEE89 pKa = 4.0AGAVVTISDD98 pKa = 3.9GSTVLGSVTAGAGGAWSFTTSALSNGTHH126 pKa = 6.82PLSVTVKK133 pKa = 9.97DD134 pKa = 3.39AAGNVSPATAATVIVDD150 pKa = 4.11TVAPTASTLTITADD164 pKa = 3.12NVTPNVTVPSGGSTNDD180 pKa = 3.29TTPTLSGTAEE190 pKa = 4.0IGSKK194 pKa = 7.88VTILDD199 pKa = 3.58GSTVLGTLTVGGSGTWSFTTATLSNGTHH227 pKa = 6.75PLSVTVTDD235 pKa = 4.04LAGNVSGTTSASVIIDD251 pKa = 3.71TVAPAAVTGLAASNNNGGSAVAIPNNGTTNDD282 pKa = 3.75NTPLLSGTAEE292 pKa = 4.01AGAKK296 pKa = 9.69ISIYY300 pKa = 10.87DD301 pKa = 3.58GATLLGTTTAASSGAWSFTTSTLTEE326 pKa = 4.22GTHH329 pKa = 5.25TLNVTATDD337 pKa = 3.4AAGNVSPNASVTLTVDD353 pKa = 3.5SVAPTNSTLTVTNDD367 pKa = 2.97SVTPNVTVPSGGYY380 pKa = 8.02TRR382 pKa = 11.84DD383 pKa = 3.38TTPVLTGTAEE393 pKa = 3.79VGSRR397 pKa = 11.84VTIYY401 pKa = 10.93DD402 pKa = 3.33GATILGSVTVGAGGTWSFTTAVLANGSHH430 pKa = 6.92PLSVTVTDD438 pKa = 3.57AAGNVSGITSSTVNVDD454 pKa = 3.42TVAPTAATALAVNTAGTTLTGTGEE478 pKa = 4.3AGTTVTVKK486 pKa = 10.73DD487 pKa = 3.75AGGNTIGTGTVGVTGSFSVTLTTAQTTGATLSVSLTDD524 pKa = 3.63AAGNTSPTSTVLGAIKK540 pKa = 10.02IVAVNDD546 pKa = 3.93LNEE549 pKa = 4.61LDD551 pKa = 4.01YY552 pKa = 10.92STTSGTITNTTGTTNTGTALLSVNLGSVLGVGVLSSSNAYY592 pKa = 8.25MFNVGTGDD600 pKa = 3.28TRR602 pKa = 11.84TVTLHH607 pKa = 6.73ASEE610 pKa = 4.69TSLISLIGSYY620 pKa = 10.5SLYY623 pKa = 10.47LYY625 pKa = 9.2QQQTDD630 pKa = 4.17GTWKK634 pKa = 10.68VISTTTNYY642 pKa = 10.01ISTTLLSLLTQTGSNITYY660 pKa = 10.69SNLGTGNYY668 pKa = 10.02AVVLGASSGISVLPSTSVTTVSDD691 pKa = 3.1ITTIAATVAATVTGNLLTNDD711 pKa = 3.65TSSVAGTVPTGTAVTSVSGHH731 pKa = 4.76TVGTSTSFSTAYY743 pKa = 8.38GTLTIDD749 pKa = 2.94SHH751 pKa = 7.59GNYY754 pKa = 9.57TYY756 pKa = 10.06TLKK759 pKa = 10.98AGLDD763 pKa = 3.47AGTLPVADD771 pKa = 3.86TFSYY775 pKa = 11.02SVTDD779 pKa = 3.57AQGVVTTATLTIDD792 pKa = 4.09LVHH795 pKa = 6.33GTTTASLLSVNSLFAEE811 pKa = 4.41ATTTTDD817 pKa = 2.73SDD819 pKa = 3.95HH820 pKa = 6.77SVSGSIWADD829 pKa = 2.66SSTSHH834 pKa = 6.68TGTLTITNEE843 pKa = 4.38HH844 pKa = 6.82GDD846 pKa = 3.74STT848 pKa = 4.46
Molecular weight: 82.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X1EQF3|A0A1X1EQF3_PANCY Proton/glutamate-aspartate symporter OS=Pantoea cypripedii OX=55209 GN=gltP PE=3 SV=1
MM1 pKa = 7.57EE2 pKa = 5.42MGDD5 pKa = 3.67FSPRR9 pKa = 11.84QQVSPAVRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84AQQLFRR25 pKa = 11.84AACCGQKK32 pKa = 9.99VFSRR36 pKa = 11.84LTMNGYY42 pKa = 10.41LKK44 pKa = 10.15IDD46 pKa = 3.45VGPFWRR52 pKa = 11.84ILSKK56 pKa = 11.11DD57 pKa = 3.41GGRR60 pKa = 11.84QWWLMDD66 pKa = 3.11HH67 pKa = 5.66EE68 pKa = 4.92TYY70 pKa = 10.66NRR72 pKa = 11.84EE73 pKa = 3.48IRR75 pKa = 11.84RR76 pKa = 3.75
MM1 pKa = 7.57EE2 pKa = 5.42MGDD5 pKa = 3.67FSPRR9 pKa = 11.84QQVSPAVRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84AQQLFRR25 pKa = 11.84AACCGQKK32 pKa = 9.99VFSRR36 pKa = 11.84LTMNGYY42 pKa = 10.41LKK44 pKa = 10.15IDD46 pKa = 3.45VGPFWRR52 pKa = 11.84ILSKK56 pKa = 11.11DD57 pKa = 3.41GGRR60 pKa = 11.84QWWLMDD66 pKa = 3.11HH67 pKa = 5.66EE68 pKa = 4.92TYY70 pKa = 10.66NRR72 pKa = 11.84EE73 pKa = 3.48IRR75 pKa = 11.84RR76 pKa = 3.75
Molecular weight: 9.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1873892 |
29 |
3919 |
320.4 |
35.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.238 ± 0.043 | 1.001 ± 0.011 |
5.202 ± 0.026 | 5.297 ± 0.032 |
3.815 ± 0.021 | 7.419 ± 0.036 |
2.329 ± 0.016 | 5.575 ± 0.028 |
3.78 ± 0.03 | 11.118 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.691 ± 0.017 | 3.603 ± 0.025 |
4.591 ± 0.022 | 4.92 ± 0.035 |
5.791 ± 0.033 | 6.119 ± 0.027 |
5.336 ± 0.036 | 6.954 ± 0.027 |
1.55 ± 0.013 | 2.67 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
