
Ruaniaceae bacterium KH17
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Ruaniaceae; unclassified Ruaniaceae
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
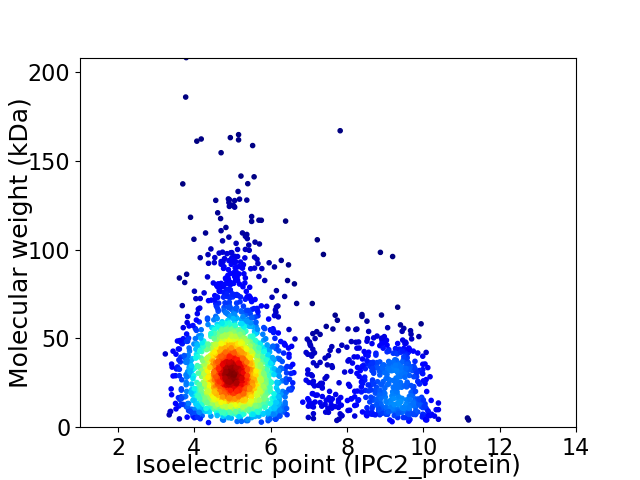
Virtual 2D-PAGE plot for 2522 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
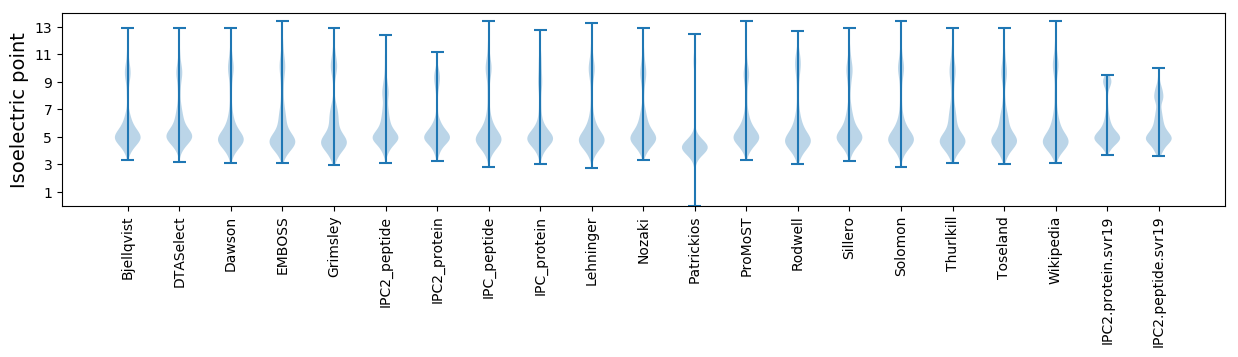
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A239QVN1|A0A239QVN1_9MICO Integrase core domain-containing protein OS=Ruaniaceae bacterium KH17 OX=1945888 GN=SAMN06298212_14719 PE=4 SV=1
MM1 pKa = 7.53KK2 pKa = 10.21FLRR5 pKa = 11.84AATALAAAALVLTACGSSIPDD26 pKa = 3.71KK27 pKa = 10.41EE28 pKa = 4.39WPTRR32 pKa = 11.84PSTDD36 pKa = 3.02GGTSTDD42 pKa = 4.27DD43 pKa = 3.46ATSSDD48 pKa = 3.95DD49 pKa = 4.6GGIFDD54 pKa = 5.4PLAGTDD60 pKa = 3.28SALAEE65 pKa = 4.59FYY67 pKa = 11.02GQDD70 pKa = 4.42VDD72 pKa = 3.86WQSCGSNLEE81 pKa = 4.28CADD84 pKa = 3.48VVVPLDD90 pKa = 3.67YY91 pKa = 11.36ANPDD95 pKa = 3.26DD96 pKa = 4.0GRR98 pKa = 11.84TVRR101 pKa = 11.84LHH103 pKa = 6.04VGVLGTAGSNSPGLFLNPGGPGGSGFAMLDD133 pKa = 3.59WIEE136 pKa = 4.22HH137 pKa = 5.96SISGSVTSTYY147 pKa = 11.18NLVGFDD153 pKa = 3.55PRR155 pKa = 11.84GVNTSDD161 pKa = 4.15AVSCLSDD168 pKa = 3.58QEE170 pKa = 3.9RR171 pKa = 11.84DD172 pKa = 3.03EE173 pKa = 4.43WNEE176 pKa = 3.74KK177 pKa = 10.65DD178 pKa = 3.4WDD180 pKa = 4.09LEE182 pKa = 4.26TATGFDD188 pKa = 5.0EE189 pKa = 4.2MTADD193 pKa = 3.87YY194 pKa = 10.78KK195 pKa = 10.93WFVDD199 pKa = 3.35KK200 pKa = 11.03CVEE203 pKa = 4.08NTPSDD208 pKa = 3.82LLEE211 pKa = 5.45FIDD214 pKa = 3.92TDD216 pKa = 3.77SAARR220 pKa = 11.84DD221 pKa = 3.62LDD223 pKa = 3.32ILRR226 pKa = 11.84AVVGQQDD233 pKa = 3.53TLDD236 pKa = 3.72YY237 pKa = 11.07LGYY240 pKa = 10.56SYY242 pKa = 10.66GTFLGAQYY250 pKa = 11.52AEE252 pKa = 4.58LFPEE256 pKa = 4.05RR257 pKa = 11.84VGAFVLDD264 pKa = 4.13GALDD268 pKa = 3.91PTLSMQDD275 pKa = 3.1MGKK278 pKa = 8.96EE279 pKa = 3.64QTLGFEE285 pKa = 4.33KK286 pKa = 10.57AIRR289 pKa = 11.84AYY291 pKa = 10.25MEE293 pKa = 4.6DD294 pKa = 3.68CLAGVGCPFSGSVEE308 pKa = 4.11NGLDD312 pKa = 3.41QLNTFFDD319 pKa = 4.16MVTATPLPTSDD330 pKa = 3.99PNRR333 pKa = 11.84DD334 pKa = 3.48LGGSGAISAVLIGLYY349 pKa = 10.04EE350 pKa = 4.13SSLWGEE356 pKa = 3.93ISNALDD362 pKa = 3.18VAMNDD367 pKa = 3.53GDD369 pKa = 4.84GSALQYY375 pKa = 9.38WADD378 pKa = 3.55VSVSRR383 pKa = 11.84LDD385 pKa = 3.89DD386 pKa = 3.37GTYY389 pKa = 10.74EE390 pKa = 4.75DD391 pKa = 4.33NSADD395 pKa = 3.33AFLAITCLDD404 pKa = 3.75YY405 pKa = 11.15PVVGNAADD413 pKa = 3.29WKK415 pKa = 10.7AAAEE419 pKa = 4.1EE420 pKa = 4.24MKK422 pKa = 10.47EE423 pKa = 4.45IAPFWQDD430 pKa = 2.47SFSYY434 pKa = 10.67TEE436 pKa = 4.64VICSQWPHH444 pKa = 4.94EE445 pKa = 4.48AEE447 pKa = 4.0RR448 pKa = 11.84TPAPVTAAGSAPILVIGTTGDD469 pKa = 3.36PATPYY474 pKa = 9.1EE475 pKa = 4.0WSEE478 pKa = 3.91ALVSQLEE485 pKa = 4.2NGHH488 pKa = 6.81LLTFEE493 pKa = 4.89GNGHH497 pKa = 4.95TAYY500 pKa = 10.26GRR502 pKa = 11.84SNSCIQNAVDD512 pKa = 5.35DD513 pKa = 4.31FLLNGTIPAEE523 pKa = 4.23GTTCC527 pKa = 4.29
MM1 pKa = 7.53KK2 pKa = 10.21FLRR5 pKa = 11.84AATALAAAALVLTACGSSIPDD26 pKa = 3.71KK27 pKa = 10.41EE28 pKa = 4.39WPTRR32 pKa = 11.84PSTDD36 pKa = 3.02GGTSTDD42 pKa = 4.27DD43 pKa = 3.46ATSSDD48 pKa = 3.95DD49 pKa = 4.6GGIFDD54 pKa = 5.4PLAGTDD60 pKa = 3.28SALAEE65 pKa = 4.59FYY67 pKa = 11.02GQDD70 pKa = 4.42VDD72 pKa = 3.86WQSCGSNLEE81 pKa = 4.28CADD84 pKa = 3.48VVVPLDD90 pKa = 3.67YY91 pKa = 11.36ANPDD95 pKa = 3.26DD96 pKa = 4.0GRR98 pKa = 11.84TVRR101 pKa = 11.84LHH103 pKa = 6.04VGVLGTAGSNSPGLFLNPGGPGGSGFAMLDD133 pKa = 3.59WIEE136 pKa = 4.22HH137 pKa = 5.96SISGSVTSTYY147 pKa = 11.18NLVGFDD153 pKa = 3.55PRR155 pKa = 11.84GVNTSDD161 pKa = 4.15AVSCLSDD168 pKa = 3.58QEE170 pKa = 3.9RR171 pKa = 11.84DD172 pKa = 3.03EE173 pKa = 4.43WNEE176 pKa = 3.74KK177 pKa = 10.65DD178 pKa = 3.4WDD180 pKa = 4.09LEE182 pKa = 4.26TATGFDD188 pKa = 5.0EE189 pKa = 4.2MTADD193 pKa = 3.87YY194 pKa = 10.78KK195 pKa = 10.93WFVDD199 pKa = 3.35KK200 pKa = 11.03CVEE203 pKa = 4.08NTPSDD208 pKa = 3.82LLEE211 pKa = 5.45FIDD214 pKa = 3.92TDD216 pKa = 3.77SAARR220 pKa = 11.84DD221 pKa = 3.62LDD223 pKa = 3.32ILRR226 pKa = 11.84AVVGQQDD233 pKa = 3.53TLDD236 pKa = 3.72YY237 pKa = 11.07LGYY240 pKa = 10.56SYY242 pKa = 10.66GTFLGAQYY250 pKa = 11.52AEE252 pKa = 4.58LFPEE256 pKa = 4.05RR257 pKa = 11.84VGAFVLDD264 pKa = 4.13GALDD268 pKa = 3.91PTLSMQDD275 pKa = 3.1MGKK278 pKa = 8.96EE279 pKa = 3.64QTLGFEE285 pKa = 4.33KK286 pKa = 10.57AIRR289 pKa = 11.84AYY291 pKa = 10.25MEE293 pKa = 4.6DD294 pKa = 3.68CLAGVGCPFSGSVEE308 pKa = 4.11NGLDD312 pKa = 3.41QLNTFFDD319 pKa = 4.16MVTATPLPTSDD330 pKa = 3.99PNRR333 pKa = 11.84DD334 pKa = 3.48LGGSGAISAVLIGLYY349 pKa = 10.04EE350 pKa = 4.13SSLWGEE356 pKa = 3.93ISNALDD362 pKa = 3.18VAMNDD367 pKa = 3.53GDD369 pKa = 4.84GSALQYY375 pKa = 9.38WADD378 pKa = 3.55VSVSRR383 pKa = 11.84LDD385 pKa = 3.89DD386 pKa = 3.37GTYY389 pKa = 10.74EE390 pKa = 4.75DD391 pKa = 4.33NSADD395 pKa = 3.33AFLAITCLDD404 pKa = 3.75YY405 pKa = 11.15PVVGNAADD413 pKa = 3.29WKK415 pKa = 10.7AAAEE419 pKa = 4.1EE420 pKa = 4.24MKK422 pKa = 10.47EE423 pKa = 4.45IAPFWQDD430 pKa = 2.47SFSYY434 pKa = 10.67TEE436 pKa = 4.64VICSQWPHH444 pKa = 4.94EE445 pKa = 4.48AEE447 pKa = 4.0RR448 pKa = 11.84TPAPVTAAGSAPILVIGTTGDD469 pKa = 3.36PATPYY474 pKa = 9.1EE475 pKa = 4.0WSEE478 pKa = 3.91ALVSQLEE485 pKa = 4.2NGHH488 pKa = 6.81LLTFEE493 pKa = 4.89GNGHH497 pKa = 4.95TAYY500 pKa = 10.26GRR502 pKa = 11.84SNSCIQNAVDD512 pKa = 5.35DD513 pKa = 4.31FLLNGTIPAEE523 pKa = 4.23GTTCC527 pKa = 4.29
Molecular weight: 56.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A239QTP6|A0A239QTP6_9MICO Cytochrome c-type biogenesis protein OS=Ruaniaceae bacterium KH17 OX=1945888 GN=SAMN06298212_12137 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.38KK7 pKa = 9.34RR8 pKa = 11.84RR9 pKa = 11.84ARR11 pKa = 11.84MAKK14 pKa = 8.94KK15 pKa = 9.75KK16 pKa = 9.66HH17 pKa = 5.58RR18 pKa = 11.84KK19 pKa = 8.32RR20 pKa = 11.84LRR22 pKa = 11.84KK23 pKa = 7.82TRR25 pKa = 11.84HH26 pKa = 3.36QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.38KK7 pKa = 9.34RR8 pKa = 11.84RR9 pKa = 11.84ARR11 pKa = 11.84MAKK14 pKa = 8.94KK15 pKa = 9.75KK16 pKa = 9.66HH17 pKa = 5.58RR18 pKa = 11.84KK19 pKa = 8.32RR20 pKa = 11.84LRR22 pKa = 11.84KK23 pKa = 7.82TRR25 pKa = 11.84HH26 pKa = 3.36QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
819933 |
24 |
2028 |
325.1 |
35.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.443 ± 0.06 | 0.611 ± 0.015 |
5.915 ± 0.045 | 6.434 ± 0.05 |
3.17 ± 0.033 | 8.634 ± 0.04 |
2.009 ± 0.02 | 4.995 ± 0.036 |
2.352 ± 0.037 | 9.988 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.112 ± 0.021 | 2.397 ± 0.025 |
5.249 ± 0.035 | 2.9 ± 0.026 |
6.684 ± 0.049 | 5.778 ± 0.032 |
6.079 ± 0.042 | 8.544 ± 0.045 |
1.515 ± 0.022 | 2.191 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
