
Brevibacterium sp. CS2
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Brevibacteriaceae; Brevibacterium; unclassified Brevibacterium
Average proteome isoelectric point is 5.94
Get precalculated fractions of proteins
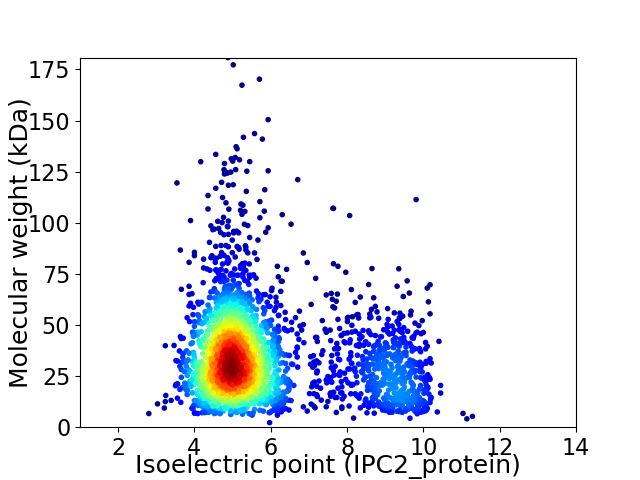
Virtual 2D-PAGE plot for 2708 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8H870|A0A4P8H870_9MICO Uncharacterized protein OS=Brevibacterium sp. CS2 OX=2575923 GN=FDF13_02840 PE=4 SV=1
MM1 pKa = 7.73KK2 pKa = 9.1LTTMSKK8 pKa = 10.24VGGVLAASVLALTACGVSNEE28 pKa = 4.46GGNSGGGGDD37 pKa = 3.55EE38 pKa = 4.11TAAGGGSEE46 pKa = 4.77VDD48 pKa = 3.77WSDD51 pKa = 3.43CTPGEE56 pKa = 4.06EE57 pKa = 4.63SADD60 pKa = 3.91TSSMEE65 pKa = 4.46ADD67 pKa = 3.41SDD69 pKa = 3.84TDD71 pKa = 3.35ITIGAFDD78 pKa = 3.56GWDD81 pKa = 3.32EE82 pKa = 4.36SFAAAHH88 pKa = 6.68LYY90 pKa = 10.26KK91 pKa = 10.79YY92 pKa = 10.75LLEE95 pKa = 4.24QDD97 pKa = 4.74GYY99 pKa = 8.53TADD102 pKa = 3.61VQSFQAGPGYY112 pKa = 9.5TGLAEE117 pKa = 4.24GDD119 pKa = 3.05IDD121 pKa = 5.04VILDD125 pKa = 3.4GWLPLTHH132 pKa = 7.31ADD134 pKa = 3.49YY135 pKa = 11.29LEE137 pKa = 4.28QYY139 pKa = 10.86GEE141 pKa = 4.7DD142 pKa = 4.14IEE144 pKa = 6.04AHH146 pKa = 5.65GCWYY150 pKa = 10.47DD151 pKa = 3.62NAKK154 pKa = 8.84LTLAVNEE161 pKa = 4.71DD162 pKa = 3.79SPAQSIADD170 pKa = 3.95LAEE173 pKa = 4.21MGDD176 pKa = 3.73EE177 pKa = 4.05YY178 pKa = 11.57GNRR181 pKa = 11.84IVGIEE186 pKa = 3.93AGAGLTKK193 pKa = 8.78TTEE196 pKa = 3.73EE197 pKa = 4.41SAIPTYY203 pKa = 10.83GLEE206 pKa = 4.24DD207 pKa = 3.56YY208 pKa = 10.76EE209 pKa = 5.44FIVSSTPAMLSEE221 pKa = 4.58LKK223 pKa = 10.53KK224 pKa = 10.7ATDD227 pKa = 3.27SGDD230 pKa = 3.85NILVTLWRR238 pKa = 11.84PHH240 pKa = 4.95WAYY243 pKa = 10.89DD244 pKa = 3.49AFPVRR249 pKa = 11.84DD250 pKa = 3.69LEE252 pKa = 4.43DD253 pKa = 3.97PEE255 pKa = 4.82GAMGEE260 pKa = 4.3AEE262 pKa = 4.81LIYY265 pKa = 10.83NFTRR269 pKa = 11.84SGFSDD274 pKa = 3.66DD275 pKa = 4.15HH276 pKa = 6.55PKK278 pKa = 10.72AGQLLKK284 pKa = 11.11NLVIDD289 pKa = 5.06DD290 pKa = 4.32EE291 pKa = 4.75ALSSLEE297 pKa = 3.39NVMFSEE303 pKa = 4.33EE304 pKa = 4.19NYY306 pKa = 10.7NGEE309 pKa = 4.32DD310 pKa = 3.27LDD312 pKa = 4.26GAVEE316 pKa = 4.34EE317 pKa = 4.53WAGDD321 pKa = 3.38NAEE324 pKa = 5.17FFEE327 pKa = 4.27NWKK330 pKa = 9.48SGSMAGG336 pKa = 3.84
MM1 pKa = 7.73KK2 pKa = 9.1LTTMSKK8 pKa = 10.24VGGVLAASVLALTACGVSNEE28 pKa = 4.46GGNSGGGGDD37 pKa = 3.55EE38 pKa = 4.11TAAGGGSEE46 pKa = 4.77VDD48 pKa = 3.77WSDD51 pKa = 3.43CTPGEE56 pKa = 4.06EE57 pKa = 4.63SADD60 pKa = 3.91TSSMEE65 pKa = 4.46ADD67 pKa = 3.41SDD69 pKa = 3.84TDD71 pKa = 3.35ITIGAFDD78 pKa = 3.56GWDD81 pKa = 3.32EE82 pKa = 4.36SFAAAHH88 pKa = 6.68LYY90 pKa = 10.26KK91 pKa = 10.79YY92 pKa = 10.75LLEE95 pKa = 4.24QDD97 pKa = 4.74GYY99 pKa = 8.53TADD102 pKa = 3.61VQSFQAGPGYY112 pKa = 9.5TGLAEE117 pKa = 4.24GDD119 pKa = 3.05IDD121 pKa = 5.04VILDD125 pKa = 3.4GWLPLTHH132 pKa = 7.31ADD134 pKa = 3.49YY135 pKa = 11.29LEE137 pKa = 4.28QYY139 pKa = 10.86GEE141 pKa = 4.7DD142 pKa = 4.14IEE144 pKa = 6.04AHH146 pKa = 5.65GCWYY150 pKa = 10.47DD151 pKa = 3.62NAKK154 pKa = 8.84LTLAVNEE161 pKa = 4.71DD162 pKa = 3.79SPAQSIADD170 pKa = 3.95LAEE173 pKa = 4.21MGDD176 pKa = 3.73EE177 pKa = 4.05YY178 pKa = 11.57GNRR181 pKa = 11.84IVGIEE186 pKa = 3.93AGAGLTKK193 pKa = 8.78TTEE196 pKa = 3.73EE197 pKa = 4.41SAIPTYY203 pKa = 10.83GLEE206 pKa = 4.24DD207 pKa = 3.56YY208 pKa = 10.76EE209 pKa = 5.44FIVSSTPAMLSEE221 pKa = 4.58LKK223 pKa = 10.53KK224 pKa = 10.7ATDD227 pKa = 3.27SGDD230 pKa = 3.85NILVTLWRR238 pKa = 11.84PHH240 pKa = 4.95WAYY243 pKa = 10.89DD244 pKa = 3.49AFPVRR249 pKa = 11.84DD250 pKa = 3.69LEE252 pKa = 4.43DD253 pKa = 3.97PEE255 pKa = 4.82GAMGEE260 pKa = 4.3AEE262 pKa = 4.81LIYY265 pKa = 10.83NFTRR269 pKa = 11.84SGFSDD274 pKa = 3.66DD275 pKa = 4.15HH276 pKa = 6.55PKK278 pKa = 10.72AGQLLKK284 pKa = 11.11NLVIDD289 pKa = 5.06DD290 pKa = 4.32EE291 pKa = 4.75ALSSLEE297 pKa = 3.39NVMFSEE303 pKa = 4.33EE304 pKa = 4.19NYY306 pKa = 10.7NGEE309 pKa = 4.32DD310 pKa = 3.27LDD312 pKa = 4.26GAVEE316 pKa = 4.34EE317 pKa = 4.53WAGDD321 pKa = 3.38NAEE324 pKa = 5.17FFEE327 pKa = 4.27NWKK330 pKa = 9.48SGSMAGG336 pKa = 3.84
Molecular weight: 35.84 kDa
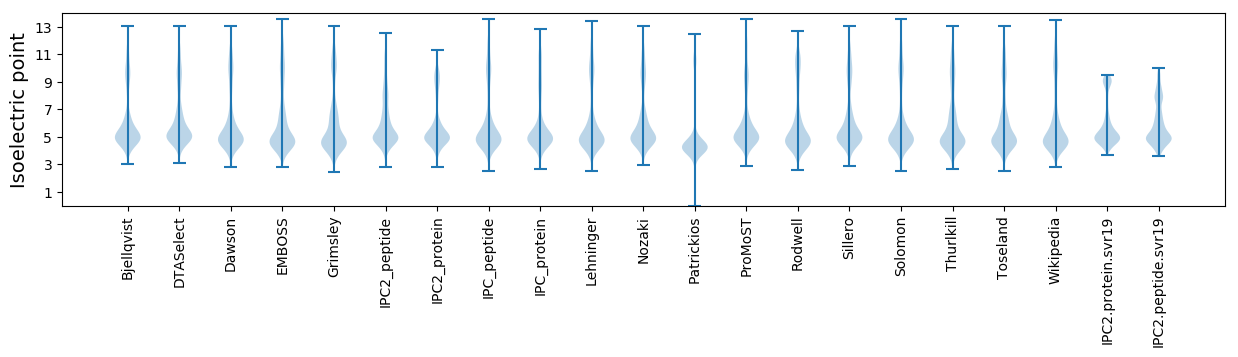
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8H6Y9|A0A4P8H6Y9_9MICO D-alanyl-D-alanine carboxypeptidase OS=Brevibacterium sp. CS2 OX=2575923 GN=FDF13_00195 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84KK12 pKa = 8.98RR13 pKa = 11.84AKK15 pKa = 8.46THH17 pKa = 5.11GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILASRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.36GRR40 pKa = 11.84NALSAA45 pKa = 3.9
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84KK12 pKa = 8.98RR13 pKa = 11.84AKK15 pKa = 8.46THH17 pKa = 5.11GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILASRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.36GRR40 pKa = 11.84NALSAA45 pKa = 3.9
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
913174 |
21 |
1651 |
337.2 |
36.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.382 ± 0.071 | 0.659 ± 0.012 |
6.202 ± 0.05 | 6.279 ± 0.049 |
3.007 ± 0.028 | 9.22 ± 0.044 |
2.233 ± 0.021 | 4.424 ± 0.029 |
1.956 ± 0.033 | 9.756 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.903 ± 0.02 | 1.81 ± 0.022 |
5.741 ± 0.04 | 2.718 ± 0.028 |
7.389 ± 0.058 | 5.568 ± 0.031 |
6.197 ± 0.035 | 8.383 ± 0.047 |
1.31 ± 0.017 | 1.863 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
