
Pseudomonas phage phi3
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Peduovirinae; Phitrevirus; Pseudomonas virus phi3
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
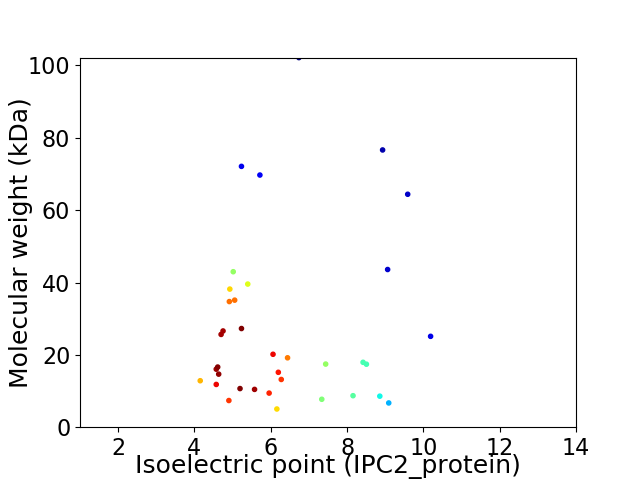
Virtual 2D-PAGE plot for 36 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
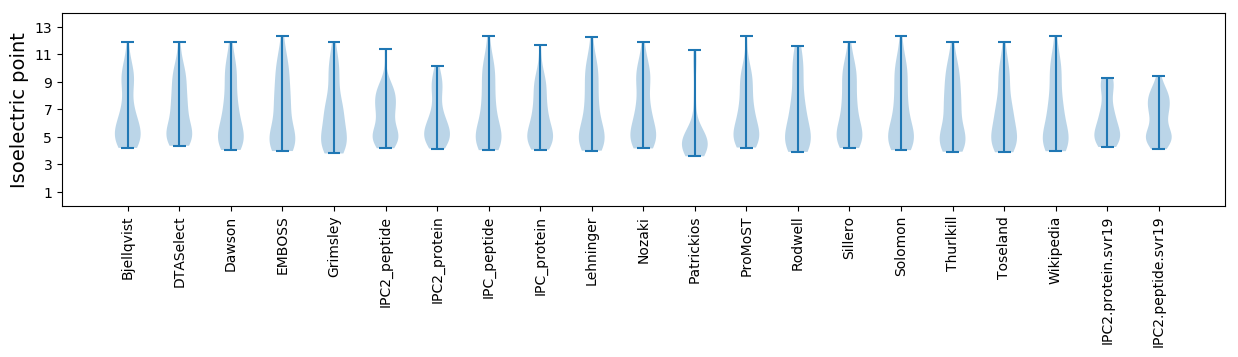
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0U4JP67|A0A0U4JP67_9CAUD Lysozyme OS=Pseudomonas phage phi3 OX=1754217 PE=3 SV=1
MM1 pKa = 7.4NEE3 pKa = 4.24SISAAMAKK11 pKa = 9.79IYY13 pKa = 10.29AYY15 pKa = 10.26DD16 pKa = 3.44QEE18 pKa = 4.66RR19 pKa = 11.84PALIAAGNQALVRR32 pKa = 11.84LAPIALQPTDD42 pKa = 3.26QARR45 pKa = 11.84TIGRR49 pKa = 11.84FLLGLYY55 pKa = 9.97NGEE58 pKa = 4.36EE59 pKa = 4.37FPFDD63 pKa = 3.55LTDD66 pKa = 4.97LRR68 pKa = 11.84GLDD71 pKa = 3.58LDD73 pKa = 5.38LFQDD77 pKa = 4.25CLAVLVMDD85 pKa = 4.55YY86 pKa = 10.99SPEE89 pKa = 4.24LEE91 pKa = 4.09VHH93 pKa = 5.94EE94 pKa = 4.79RR95 pKa = 11.84VPDD98 pKa = 4.05GDD100 pKa = 5.59AIWQRR105 pKa = 11.84LIEE108 pKa = 4.29LWAPEE113 pKa = 3.86AAA115 pKa = 4.45
MM1 pKa = 7.4NEE3 pKa = 4.24SISAAMAKK11 pKa = 9.79IYY13 pKa = 10.29AYY15 pKa = 10.26DD16 pKa = 3.44QEE18 pKa = 4.66RR19 pKa = 11.84PALIAAGNQALVRR32 pKa = 11.84LAPIALQPTDD42 pKa = 3.26QARR45 pKa = 11.84TIGRR49 pKa = 11.84FLLGLYY55 pKa = 9.97NGEE58 pKa = 4.36EE59 pKa = 4.37FPFDD63 pKa = 3.55LTDD66 pKa = 4.97LRR68 pKa = 11.84GLDD71 pKa = 3.58LDD73 pKa = 5.38LFQDD77 pKa = 4.25CLAVLVMDD85 pKa = 4.55YY86 pKa = 10.99SPEE89 pKa = 4.24LEE91 pKa = 4.09VHH93 pKa = 5.94EE94 pKa = 4.79RR95 pKa = 11.84VPDD98 pKa = 4.05GDD100 pKa = 5.59AIWQRR105 pKa = 11.84LIEE108 pKa = 4.29LWAPEE113 pKa = 3.86AAA115 pKa = 4.45
Molecular weight: 12.85 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0U4J933|A0A0U4J933_9CAUD Putative head completion/stabilization protein OS=Pseudomonas phage phi3 OX=1754217 PE=4 SV=1
MM1 pKa = 7.68KK2 pKa = 9.91EE3 pKa = 4.17LSPSAGSALSGAPQQWHH20 pKa = 7.39DD21 pKa = 3.73IDD23 pKa = 3.24WCRR26 pKa = 11.84VQRR29 pKa = 11.84NVRR32 pKa = 11.84GMQVRR37 pKa = 11.84IAKK40 pKa = 9.92ACRR43 pKa = 11.84EE44 pKa = 4.33GKK46 pKa = 8.2WRR48 pKa = 11.84RR49 pKa = 11.84VKK51 pKa = 10.59ALQRR55 pKa = 11.84MLTRR59 pKa = 11.84STSARR64 pKa = 11.84YY65 pKa = 8.6LAVRR69 pKa = 11.84RR70 pKa = 11.84VTEE73 pKa = 4.01NQGKK77 pKa = 7.83RR78 pKa = 11.84TAGVDD83 pKa = 3.05RR84 pKa = 11.84VLWDD88 pKa = 3.67TPDD91 pKa = 3.98TKK93 pKa = 10.42WKK95 pKa = 10.02AAQGLKK101 pKa = 10.15RR102 pKa = 11.84RR103 pKa = 11.84GYY105 pKa = 8.19QPRR108 pKa = 11.84PLRR111 pKa = 11.84RR112 pKa = 11.84VFIPKK117 pKa = 10.18SNGKK121 pKa = 8.54EE122 pKa = 3.8RR123 pKa = 11.84PLGIPTMTDD132 pKa = 2.45RR133 pKa = 11.84AMQALYY139 pKa = 10.88LLALSPIAEE148 pKa = 4.42TTGDD152 pKa = 3.65PNSYY156 pKa = 9.88GFRR159 pKa = 11.84IEE161 pKa = 5.13RR162 pKa = 11.84STADD166 pKa = 3.07AMSQLFVCLSGKK178 pKa = 10.37ASAQWILEE186 pKa = 3.87ADD188 pKa = 3.15IQGCFDD194 pKa = 5.61HH195 pKa = 7.37INHH198 pKa = 7.1DD199 pKa = 3.34WLLNHH204 pKa = 6.0VPTDD208 pKa = 3.72KK209 pKa = 11.18VILRR213 pKa = 11.84KK214 pKa = 8.71WLKK217 pKa = 10.56AGVIHH222 pKa = 7.52KK223 pKa = 9.9GQLQATDD230 pKa = 3.25AGTPQGGIISPTLANMVLDD249 pKa = 4.09GLEE252 pKa = 4.27SQLKK256 pKa = 8.89RR257 pKa = 11.84HH258 pKa = 6.22LGVTRR263 pKa = 11.84AKK265 pKa = 10.24KK266 pKa = 10.41LKK268 pKa = 10.41LNVVRR273 pKa = 11.84YY274 pKa = 10.11ADD276 pKa = 3.94DD277 pKa = 3.83FVITGVSPEE286 pKa = 4.03VLEE289 pKa = 5.14KK290 pKa = 10.49EE291 pKa = 4.23VKK293 pKa = 9.34PWVEE297 pKa = 3.21QFLAVRR303 pKa = 11.84GLQLSLEE310 pKa = 4.44KK311 pKa = 10.36TRR313 pKa = 11.84IAHH316 pKa = 6.61IDD318 pKa = 3.22QGFDD322 pKa = 3.21FLGWNFRR329 pKa = 11.84KK330 pKa = 10.57YY331 pKa = 10.45NGKK334 pKa = 9.27LLIKK338 pKa = 10.13PSQKK342 pKa = 9.7NAKK345 pKa = 9.59AFYY348 pKa = 9.68GKK350 pKa = 9.94VKK352 pKa = 10.36EE353 pKa = 4.31VIEE356 pKa = 4.2TSRR359 pKa = 11.84TARR362 pKa = 11.84QEE364 pKa = 3.84DD365 pKa = 5.0LIRR368 pKa = 11.84TLNPILRR375 pKa = 11.84GWALYY380 pKa = 8.2HH381 pKa = 5.69QPVVAKK387 pKa = 8.36QTFSRR392 pKa = 11.84MDD394 pKa = 2.92NRR396 pKa = 11.84VFLKK400 pKa = 9.89LWRR403 pKa = 11.84WAKK406 pKa = 9.77RR407 pKa = 11.84RR408 pKa = 11.84HH409 pKa = 5.58PNKK412 pKa = 9.91SLDD415 pKa = 3.47WIRR418 pKa = 11.84RR419 pKa = 11.84RR420 pKa = 11.84YY421 pKa = 9.46FRR423 pKa = 11.84IHH425 pKa = 7.19GDD427 pKa = 3.52KK428 pKa = 9.76TWVFATTVLEE438 pKa = 4.41SNGKK442 pKa = 8.57KK443 pKa = 9.91RR444 pKa = 11.84EE445 pKa = 3.82VALYY449 pKa = 10.19SLASTPIEE457 pKa = 3.87RR458 pKa = 11.84HH459 pKa = 5.24RR460 pKa = 11.84KK461 pKa = 9.15VSGEE465 pKa = 3.89YY466 pKa = 10.25NPYY469 pKa = 10.07DD470 pKa = 3.6PAMEE474 pKa = 5.2AIGEE478 pKa = 4.23KK479 pKa = 10.57LRR481 pKa = 11.84TEE483 pKa = 4.11RR484 pKa = 11.84MLKK487 pKa = 8.95KK488 pKa = 10.4LKK490 pKa = 10.51YY491 pKa = 8.46RR492 pKa = 11.84TQIRR496 pKa = 11.84NLYY499 pKa = 9.65LSQKK503 pKa = 9.56GLCSLCNQPITRR515 pKa = 11.84EE516 pKa = 4.32TGWHH520 pKa = 6.25DD521 pKa = 2.94HH522 pKa = 6.7HH523 pKa = 8.2ILYY526 pKa = 10.22RR527 pKa = 11.84SQGGGDD533 pKa = 3.33SLEE536 pKa = 4.07NRR538 pKa = 11.84VLLHH542 pKa = 6.26PACHH546 pKa = 5.42QQLHH550 pKa = 5.89ARR552 pKa = 11.84GLTVSKK558 pKa = 9.94PARR561 pKa = 3.67
MM1 pKa = 7.68KK2 pKa = 9.91EE3 pKa = 4.17LSPSAGSALSGAPQQWHH20 pKa = 7.39DD21 pKa = 3.73IDD23 pKa = 3.24WCRR26 pKa = 11.84VQRR29 pKa = 11.84NVRR32 pKa = 11.84GMQVRR37 pKa = 11.84IAKK40 pKa = 9.92ACRR43 pKa = 11.84EE44 pKa = 4.33GKK46 pKa = 8.2WRR48 pKa = 11.84RR49 pKa = 11.84VKK51 pKa = 10.59ALQRR55 pKa = 11.84MLTRR59 pKa = 11.84STSARR64 pKa = 11.84YY65 pKa = 8.6LAVRR69 pKa = 11.84RR70 pKa = 11.84VTEE73 pKa = 4.01NQGKK77 pKa = 7.83RR78 pKa = 11.84TAGVDD83 pKa = 3.05RR84 pKa = 11.84VLWDD88 pKa = 3.67TPDD91 pKa = 3.98TKK93 pKa = 10.42WKK95 pKa = 10.02AAQGLKK101 pKa = 10.15RR102 pKa = 11.84RR103 pKa = 11.84GYY105 pKa = 8.19QPRR108 pKa = 11.84PLRR111 pKa = 11.84RR112 pKa = 11.84VFIPKK117 pKa = 10.18SNGKK121 pKa = 8.54EE122 pKa = 3.8RR123 pKa = 11.84PLGIPTMTDD132 pKa = 2.45RR133 pKa = 11.84AMQALYY139 pKa = 10.88LLALSPIAEE148 pKa = 4.42TTGDD152 pKa = 3.65PNSYY156 pKa = 9.88GFRR159 pKa = 11.84IEE161 pKa = 5.13RR162 pKa = 11.84STADD166 pKa = 3.07AMSQLFVCLSGKK178 pKa = 10.37ASAQWILEE186 pKa = 3.87ADD188 pKa = 3.15IQGCFDD194 pKa = 5.61HH195 pKa = 7.37INHH198 pKa = 7.1DD199 pKa = 3.34WLLNHH204 pKa = 6.0VPTDD208 pKa = 3.72KK209 pKa = 11.18VILRR213 pKa = 11.84KK214 pKa = 8.71WLKK217 pKa = 10.56AGVIHH222 pKa = 7.52KK223 pKa = 9.9GQLQATDD230 pKa = 3.25AGTPQGGIISPTLANMVLDD249 pKa = 4.09GLEE252 pKa = 4.27SQLKK256 pKa = 8.89RR257 pKa = 11.84HH258 pKa = 6.22LGVTRR263 pKa = 11.84AKK265 pKa = 10.24KK266 pKa = 10.41LKK268 pKa = 10.41LNVVRR273 pKa = 11.84YY274 pKa = 10.11ADD276 pKa = 3.94DD277 pKa = 3.83FVITGVSPEE286 pKa = 4.03VLEE289 pKa = 5.14KK290 pKa = 10.49EE291 pKa = 4.23VKK293 pKa = 9.34PWVEE297 pKa = 3.21QFLAVRR303 pKa = 11.84GLQLSLEE310 pKa = 4.44KK311 pKa = 10.36TRR313 pKa = 11.84IAHH316 pKa = 6.61IDD318 pKa = 3.22QGFDD322 pKa = 3.21FLGWNFRR329 pKa = 11.84KK330 pKa = 10.57YY331 pKa = 10.45NGKK334 pKa = 9.27LLIKK338 pKa = 10.13PSQKK342 pKa = 9.7NAKK345 pKa = 9.59AFYY348 pKa = 9.68GKK350 pKa = 9.94VKK352 pKa = 10.36EE353 pKa = 4.31VIEE356 pKa = 4.2TSRR359 pKa = 11.84TARR362 pKa = 11.84QEE364 pKa = 3.84DD365 pKa = 5.0LIRR368 pKa = 11.84TLNPILRR375 pKa = 11.84GWALYY380 pKa = 8.2HH381 pKa = 5.69QPVVAKK387 pKa = 8.36QTFSRR392 pKa = 11.84MDD394 pKa = 2.92NRR396 pKa = 11.84VFLKK400 pKa = 9.89LWRR403 pKa = 11.84WAKK406 pKa = 9.77RR407 pKa = 11.84RR408 pKa = 11.84HH409 pKa = 5.58PNKK412 pKa = 9.91SLDD415 pKa = 3.47WIRR418 pKa = 11.84RR419 pKa = 11.84RR420 pKa = 11.84YY421 pKa = 9.46FRR423 pKa = 11.84IHH425 pKa = 7.19GDD427 pKa = 3.52KK428 pKa = 9.76TWVFATTVLEE438 pKa = 4.41SNGKK442 pKa = 8.57KK443 pKa = 9.91RR444 pKa = 11.84EE445 pKa = 3.82VALYY449 pKa = 10.19SLASTPIEE457 pKa = 3.87RR458 pKa = 11.84HH459 pKa = 5.24RR460 pKa = 11.84KK461 pKa = 9.15VSGEE465 pKa = 3.89YY466 pKa = 10.25NPYY469 pKa = 10.07DD470 pKa = 3.6PAMEE474 pKa = 5.2AIGEE478 pKa = 4.23KK479 pKa = 10.57LRR481 pKa = 11.84TEE483 pKa = 4.11RR484 pKa = 11.84MLKK487 pKa = 8.95KK488 pKa = 10.4LKK490 pKa = 10.51YY491 pKa = 8.46RR492 pKa = 11.84TQIRR496 pKa = 11.84NLYY499 pKa = 9.65LSQKK503 pKa = 9.56GLCSLCNQPITRR515 pKa = 11.84EE516 pKa = 4.32TGWHH520 pKa = 6.25DD521 pKa = 2.94HH522 pKa = 6.7HH523 pKa = 8.2ILYY526 pKa = 10.22RR527 pKa = 11.84SQGGGDD533 pKa = 3.33SLEE536 pKa = 4.07NRR538 pKa = 11.84VLLHH542 pKa = 6.26PACHH546 pKa = 5.42QQLHH550 pKa = 5.89ARR552 pKa = 11.84GLTVSKK558 pKa = 9.94PARR561 pKa = 3.67
Molecular weight: 64.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8948 |
46 |
896 |
248.6 |
27.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.03 ± 0.57 | 0.793 ± 0.139 |
5.767 ± 0.257 | 6.258 ± 0.346 |
3.375 ± 0.259 | 7.432 ± 0.346 |
2.09 ± 0.216 | 3.911 ± 0.26 |
4.079 ± 0.303 | 10.963 ± 0.448 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.833 ± 0.212 | 3.431 ± 0.288 |
4.973 ± 0.243 | 5.174 ± 0.315 |
7.566 ± 0.511 | 4.906 ± 0.233 |
5.331 ± 0.296 | 7.097 ± 0.322 |
1.598 ± 0.143 | 2.392 ± 0.189 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
