
Streptomyces yanglinensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
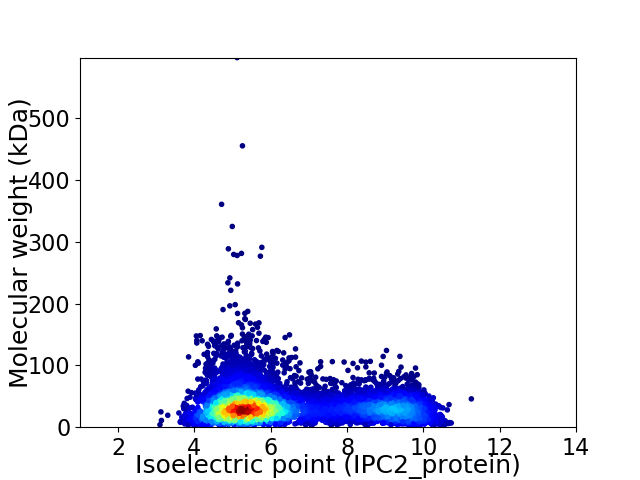
Virtual 2D-PAGE plot for 8055 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
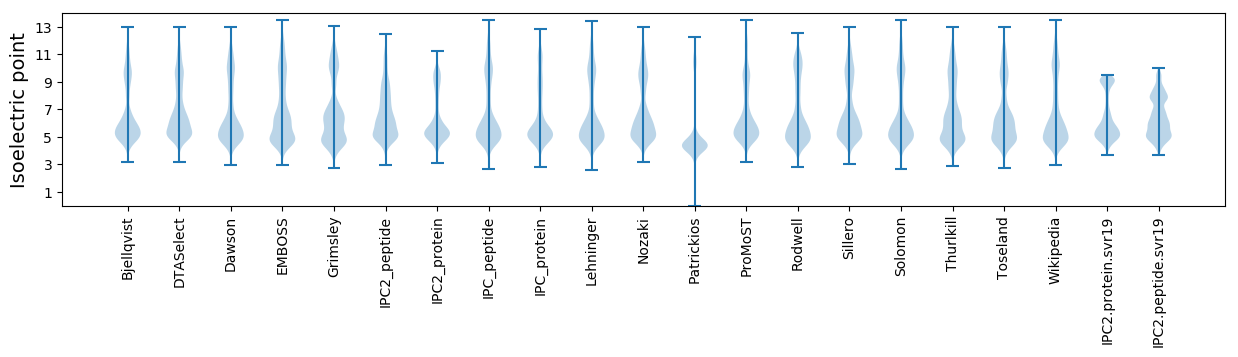
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H5Y137|A0A1H5Y137_9ACTN Protein-tyrosine phosphatase OS=Streptomyces yanglinensis OX=310779 GN=SAMN05216223_103511 PE=4 SV=1
MM1 pKa = 7.57SSHH4 pKa = 6.54FSGRR8 pKa = 11.84RR9 pKa = 11.84AFRR12 pKa = 11.84AASALTLSLGAGLAAPLLAAGSAGAATPSATAGLSDD48 pKa = 3.53NAIVYY53 pKa = 7.66TAAAGQANKK62 pKa = 8.46VTVTSSLTDD71 pKa = 3.32GNTKK75 pKa = 7.78VTYY78 pKa = 10.7VIDD81 pKa = 4.04DD82 pKa = 3.9VVPITAGDD90 pKa = 3.77GCTYY94 pKa = 7.71PTSTDD99 pKa = 3.09HH100 pKa = 6.69TKK102 pKa = 10.53VSCAAPMLDD111 pKa = 4.08TEE113 pKa = 5.03DD114 pKa = 4.2PYY116 pKa = 11.66ATLEE120 pKa = 4.09LALGDD125 pKa = 3.76GDD127 pKa = 5.63DD128 pKa = 3.88AVTYY132 pKa = 11.26ANGTNQAYY140 pKa = 9.89YY141 pKa = 10.28FARR144 pKa = 11.84FDD146 pKa = 4.32LGDD149 pKa = 3.85GKK151 pKa = 9.68DD152 pKa = 3.48TYY154 pKa = 11.22TEE156 pKa = 3.98TGKK159 pKa = 10.08VQGNDD164 pKa = 3.35VLGGAGDD171 pKa = 3.85DD172 pKa = 3.83TLTVSSYY179 pKa = 9.79TVALGGDD186 pKa = 4.06DD187 pKa = 4.64NDD189 pKa = 4.92TIHH192 pKa = 7.15AGADD196 pKa = 3.6TIVQGGNGNDD206 pKa = 3.41TVDD209 pKa = 3.43ASGAGSNVDD218 pKa = 3.27GGAGNDD224 pKa = 3.89VINGGDD230 pKa = 3.5GRR232 pKa = 11.84QDD234 pKa = 3.41LSGGDD239 pKa = 4.14GNDD242 pKa = 3.5KK243 pKa = 10.66IYY245 pKa = 10.91GGKK248 pKa = 10.4GDD250 pKa = 4.09DD251 pKa = 3.73FLYY254 pKa = 10.68GGKK257 pKa = 10.35GDD259 pKa = 5.28DD260 pKa = 3.32ILYY263 pKa = 10.65GNSGNDD269 pKa = 3.44TIYY272 pKa = 11.22GNSGNDD278 pKa = 3.47GVYY281 pKa = 10.48GGPGTDD287 pKa = 3.54TLSGGPGTNIVHH299 pKa = 6.49QDD301 pKa = 2.88
MM1 pKa = 7.57SSHH4 pKa = 6.54FSGRR8 pKa = 11.84RR9 pKa = 11.84AFRR12 pKa = 11.84AASALTLSLGAGLAAPLLAAGSAGAATPSATAGLSDD48 pKa = 3.53NAIVYY53 pKa = 7.66TAAAGQANKK62 pKa = 8.46VTVTSSLTDD71 pKa = 3.32GNTKK75 pKa = 7.78VTYY78 pKa = 10.7VIDD81 pKa = 4.04DD82 pKa = 3.9VVPITAGDD90 pKa = 3.77GCTYY94 pKa = 7.71PTSTDD99 pKa = 3.09HH100 pKa = 6.69TKK102 pKa = 10.53VSCAAPMLDD111 pKa = 4.08TEE113 pKa = 5.03DD114 pKa = 4.2PYY116 pKa = 11.66ATLEE120 pKa = 4.09LALGDD125 pKa = 3.76GDD127 pKa = 5.63DD128 pKa = 3.88AVTYY132 pKa = 11.26ANGTNQAYY140 pKa = 9.89YY141 pKa = 10.28FARR144 pKa = 11.84FDD146 pKa = 4.32LGDD149 pKa = 3.85GKK151 pKa = 9.68DD152 pKa = 3.48TYY154 pKa = 11.22TEE156 pKa = 3.98TGKK159 pKa = 10.08VQGNDD164 pKa = 3.35VLGGAGDD171 pKa = 3.85DD172 pKa = 3.83TLTVSSYY179 pKa = 9.79TVALGGDD186 pKa = 4.06DD187 pKa = 4.64NDD189 pKa = 4.92TIHH192 pKa = 7.15AGADD196 pKa = 3.6TIVQGGNGNDD206 pKa = 3.41TVDD209 pKa = 3.43ASGAGSNVDD218 pKa = 3.27GGAGNDD224 pKa = 3.89VINGGDD230 pKa = 3.5GRR232 pKa = 11.84QDD234 pKa = 3.41LSGGDD239 pKa = 4.14GNDD242 pKa = 3.5KK243 pKa = 10.66IYY245 pKa = 10.91GGKK248 pKa = 10.4GDD250 pKa = 4.09DD251 pKa = 3.73FLYY254 pKa = 10.68GGKK257 pKa = 10.35GDD259 pKa = 5.28DD260 pKa = 3.32ILYY263 pKa = 10.65GNSGNDD269 pKa = 3.44TIYY272 pKa = 11.22GNSGNDD278 pKa = 3.47GVYY281 pKa = 10.48GGPGTDD287 pKa = 3.54TLSGGPGTNIVHH299 pKa = 6.49QDD301 pKa = 2.88
Molecular weight: 29.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H6BU77|A0A1H6BU77_9ACTN Uncharacterized protein OS=Streptomyces yanglinensis OX=310779 GN=SAMN05216223_107243 PE=4 SV=1
MM1 pKa = 6.39TTVPVARR8 pKa = 11.84AVMTTAVAPATVGTTAGMTVPATTVLVMTAPATTVPAVPVVASAVMTARR57 pKa = 11.84VMTAPAVRR65 pKa = 11.84VVMTVLPGVAASAVTTVPVAVPVVGSVAMTVPAVPVVASAVMTARR110 pKa = 11.84VMTVGTTARR119 pKa = 11.84AVRR122 pKa = 11.84VVMTVLPGVAASAVTTVPVAVPVVGSVAMTVPVAVPVVGSVVTTVPAVPVVASAVTTGRR181 pKa = 11.84TGRR184 pKa = 11.84ATTVPVTIVPVAAPVSVGTTARR206 pKa = 11.84VMTARR211 pKa = 11.84VTTVRR216 pKa = 11.84AVTTTVVVSAATTAVTTVPVVRR238 pKa = 11.84AAMTTVAVSVAMTGRR253 pKa = 11.84TGRR256 pKa = 11.84ATTVPVTIVPVAAPVSVGTTARR278 pKa = 11.84VMTARR283 pKa = 11.84VTTVRR288 pKa = 11.84AVTTTVVVSAATTAVTTVPVVRR310 pKa = 11.84AAMTTVAVSVAMTGRR325 pKa = 11.84TGRR328 pKa = 11.84ATTVPVTIVPVAAPVSVGTTARR350 pKa = 11.84VMTARR355 pKa = 11.84ATTAGTSVLATTVPAGSVVTTVRR378 pKa = 11.84TGRR381 pKa = 11.84VMTVGTTARR390 pKa = 11.84AVRR393 pKa = 11.84GVMTVPPGVVASAVMTARR411 pKa = 11.84VTTVVTTGGTTVRR424 pKa = 11.84PVRR427 pKa = 11.84AGTVTGSAAAAGARR441 pKa = 11.84WCRR444 pKa = 11.84AAAGSRR450 pKa = 11.84AAAATTTAGTTGNRR464 pKa = 11.84SGGCRR469 pKa = 11.84SRR471 pKa = 11.84TTT473 pKa = 3.13
MM1 pKa = 6.39TTVPVARR8 pKa = 11.84AVMTTAVAPATVGTTAGMTVPATTVLVMTAPATTVPAVPVVASAVMTARR57 pKa = 11.84VMTAPAVRR65 pKa = 11.84VVMTVLPGVAASAVTTVPVAVPVVGSVAMTVPAVPVVASAVMTARR110 pKa = 11.84VMTVGTTARR119 pKa = 11.84AVRR122 pKa = 11.84VVMTVLPGVAASAVTTVPVAVPVVGSVAMTVPVAVPVVGSVVTTVPAVPVVASAVTTGRR181 pKa = 11.84TGRR184 pKa = 11.84ATTVPVTIVPVAAPVSVGTTARR206 pKa = 11.84VMTARR211 pKa = 11.84VTTVRR216 pKa = 11.84AVTTTVVVSAATTAVTTVPVVRR238 pKa = 11.84AAMTTVAVSVAMTGRR253 pKa = 11.84TGRR256 pKa = 11.84ATTVPVTIVPVAAPVSVGTTARR278 pKa = 11.84VMTARR283 pKa = 11.84VTTVRR288 pKa = 11.84AVTTTVVVSAATTAVTTVPVVRR310 pKa = 11.84AAMTTVAVSVAMTGRR325 pKa = 11.84TGRR328 pKa = 11.84ATTVPVTIVPVAAPVSVGTTARR350 pKa = 11.84VMTARR355 pKa = 11.84ATTAGTSVLATTVPAGSVVTTVRR378 pKa = 11.84TGRR381 pKa = 11.84VMTVGTTARR390 pKa = 11.84AVRR393 pKa = 11.84GVMTVPPGVVASAVMTARR411 pKa = 11.84VTTVVTTGGTTVRR424 pKa = 11.84PVRR427 pKa = 11.84AGTVTGSAAAAGARR441 pKa = 11.84WCRR444 pKa = 11.84AAAGSRR450 pKa = 11.84AAAATTTAGTTGNRR464 pKa = 11.84SGGCRR469 pKa = 11.84SRR471 pKa = 11.84TTT473 pKa = 3.13
Molecular weight: 45.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2771259 |
26 |
5593 |
344.0 |
36.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.194 ± 0.041 | 0.756 ± 0.008 |
6.039 ± 0.02 | 5.06 ± 0.029 |
2.67 ± 0.016 | 9.886 ± 0.03 |
2.323 ± 0.013 | 2.987 ± 0.018 |
1.777 ± 0.018 | 10.076 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.657 ± 0.01 | 1.775 ± 0.015 |
6.425 ± 0.027 | 2.821 ± 0.018 |
7.859 ± 0.035 | 5.35 ± 0.023 |
6.333 ± 0.029 | 8.398 ± 0.023 |
1.516 ± 0.011 | 2.098 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
