
Pepper golden mosaic virus-[CR]
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Begomovirus; Pepper golden mosaic virus
Average proteome isoelectric point is 8.56
Get precalculated fractions of proteins
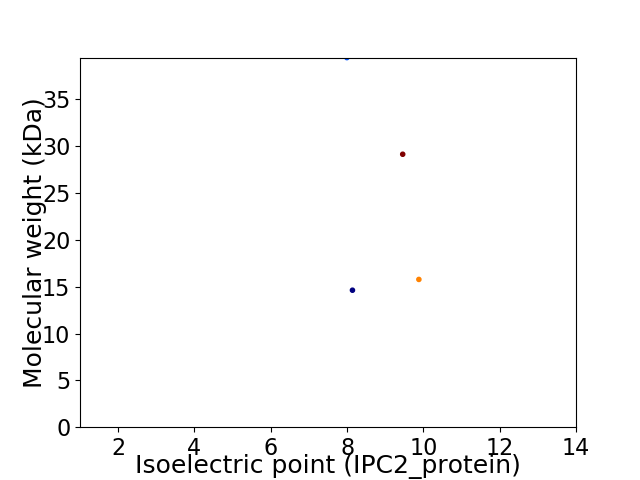
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9QJK6|Q9QJK6_9GEMI Transcriptional activator protein OS=Pepper golden mosaic virus-[CR] OX=223302 GN=AC2 PE=3 SV=1
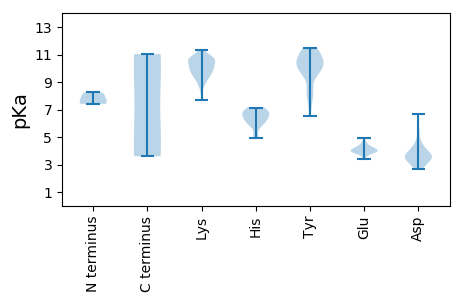
MM1 pKa = 7.93PLPPKK6 pKa = 10.4SFHH9 pKa = 6.14LQCKK13 pKa = 10.13NIFLTYY19 pKa = 8.19PQCDD23 pKa = 3.41IPKK26 pKa = 10.43DD27 pKa = 3.5EE28 pKa = 4.92ALEE31 pKa = 3.94MLQSLKK37 pKa = 10.36WSVVKK42 pKa = 10.05PIYY45 pKa = 10.04IRR47 pKa = 11.84VSRR50 pKa = 11.84EE51 pKa = 3.46EE52 pKa = 4.04HH53 pKa = 6.5SDD55 pKa = 3.62GFPHH59 pKa = 6.6LHH61 pKa = 6.91CLIQLTGKK69 pKa = 10.57CNIKK73 pKa = 10.14DD74 pKa = 3.38ARR76 pKa = 11.84FFDD79 pKa = 3.48ITHH82 pKa = 7.1PRR84 pKa = 11.84RR85 pKa = 11.84SAQFHH90 pKa = 6.88PNVQAAKK97 pKa = 9.12DD98 pKa = 3.63TNAVKK103 pKa = 10.64NYY105 pKa = 6.5ITKK108 pKa = 10.81DD109 pKa = 2.87GDD111 pKa = 3.67YY112 pKa = 11.07CEE114 pKa = 4.23TGQYY118 pKa = 9.68KK119 pKa = 10.39VSGGSKK125 pKa = 9.95ANKK128 pKa = 9.55DD129 pKa = 3.77DD130 pKa = 4.11VYY132 pKa = 11.43HH133 pKa = 6.34NAVNAASAGEE143 pKa = 3.84ALDD146 pKa = 4.26IIKK149 pKa = 10.81AGDD152 pKa = 3.56PRR154 pKa = 11.84TFIVSYY160 pKa = 11.0HH161 pKa = 4.97NVKK164 pKa = 10.87SNIEE168 pKa = 4.05RR169 pKa = 11.84LFKK172 pKa = 10.52PPPKK176 pKa = 9.79QWTPPYY182 pKa = 9.66PLSSFNNVPEE192 pKa = 4.22DD193 pKa = 3.46MQRR196 pKa = 11.84CVAEE200 pKa = 3.84YY201 pKa = 10.13FGRR204 pKa = 11.84SSAARR209 pKa = 11.84PDD211 pKa = 3.51RR212 pKa = 11.84PISIIIEE219 pKa = 4.12GDD221 pKa = 3.42SRR223 pKa = 11.84SGKK226 pKa = 7.69TMWARR231 pKa = 11.84ALGPHH236 pKa = 6.63NYY238 pKa = 10.14LSGHH242 pKa = 6.97LDD244 pKa = 3.71FNSKK248 pKa = 10.15VYY250 pKa = 11.23SNDD253 pKa = 2.67VEE255 pKa = 4.6YY256 pKa = 11.36NVIDD260 pKa = 5.95DD261 pKa = 4.05ITPHH265 pKa = 5.11YY266 pKa = 10.58LKK268 pKa = 10.62LKK270 pKa = 9.09HH271 pKa = 5.7WKK273 pKa = 9.57EE274 pKa = 4.04LIGAQRR280 pKa = 11.84DD281 pKa = 3.53WQSNCKK287 pKa = 9.41YY288 pKa = 10.21GKK290 pKa = 9.18PVQIKK295 pKa = 10.19GGIPSIVLCNPGEE308 pKa = 4.25GSSYY312 pKa = 10.53IAFLNKK318 pKa = 9.95EE319 pKa = 3.85EE320 pKa = 4.09NAPLRR325 pKa = 11.84AWTQKK330 pKa = 9.91NAQFVILHH338 pKa = 6.13APLYY342 pKa = 10.56KK343 pKa = 9.81STAQDD348 pKa = 3.02SS349 pKa = 3.81
MM1 pKa = 7.93PLPPKK6 pKa = 10.4SFHH9 pKa = 6.14LQCKK13 pKa = 10.13NIFLTYY19 pKa = 8.19PQCDD23 pKa = 3.41IPKK26 pKa = 10.43DD27 pKa = 3.5EE28 pKa = 4.92ALEE31 pKa = 3.94MLQSLKK37 pKa = 10.36WSVVKK42 pKa = 10.05PIYY45 pKa = 10.04IRR47 pKa = 11.84VSRR50 pKa = 11.84EE51 pKa = 3.46EE52 pKa = 4.04HH53 pKa = 6.5SDD55 pKa = 3.62GFPHH59 pKa = 6.6LHH61 pKa = 6.91CLIQLTGKK69 pKa = 10.57CNIKK73 pKa = 10.14DD74 pKa = 3.38ARR76 pKa = 11.84FFDD79 pKa = 3.48ITHH82 pKa = 7.1PRR84 pKa = 11.84RR85 pKa = 11.84SAQFHH90 pKa = 6.88PNVQAAKK97 pKa = 9.12DD98 pKa = 3.63TNAVKK103 pKa = 10.64NYY105 pKa = 6.5ITKK108 pKa = 10.81DD109 pKa = 2.87GDD111 pKa = 3.67YY112 pKa = 11.07CEE114 pKa = 4.23TGQYY118 pKa = 9.68KK119 pKa = 10.39VSGGSKK125 pKa = 9.95ANKK128 pKa = 9.55DD129 pKa = 3.77DD130 pKa = 4.11VYY132 pKa = 11.43HH133 pKa = 6.34NAVNAASAGEE143 pKa = 3.84ALDD146 pKa = 4.26IIKK149 pKa = 10.81AGDD152 pKa = 3.56PRR154 pKa = 11.84TFIVSYY160 pKa = 11.0HH161 pKa = 4.97NVKK164 pKa = 10.87SNIEE168 pKa = 4.05RR169 pKa = 11.84LFKK172 pKa = 10.52PPPKK176 pKa = 9.79QWTPPYY182 pKa = 9.66PLSSFNNVPEE192 pKa = 4.22DD193 pKa = 3.46MQRR196 pKa = 11.84CVAEE200 pKa = 3.84YY201 pKa = 10.13FGRR204 pKa = 11.84SSAARR209 pKa = 11.84PDD211 pKa = 3.51RR212 pKa = 11.84PISIIIEE219 pKa = 4.12GDD221 pKa = 3.42SRR223 pKa = 11.84SGKK226 pKa = 7.69TMWARR231 pKa = 11.84ALGPHH236 pKa = 6.63NYY238 pKa = 10.14LSGHH242 pKa = 6.97LDD244 pKa = 3.71FNSKK248 pKa = 10.15VYY250 pKa = 11.23SNDD253 pKa = 2.67VEE255 pKa = 4.6YY256 pKa = 11.36NVIDD260 pKa = 5.95DD261 pKa = 4.05ITPHH265 pKa = 5.11YY266 pKa = 10.58LKK268 pKa = 10.62LKK270 pKa = 9.09HH271 pKa = 5.7WKK273 pKa = 9.57EE274 pKa = 4.04LIGAQRR280 pKa = 11.84DD281 pKa = 3.53WQSNCKK287 pKa = 9.41YY288 pKa = 10.21GKK290 pKa = 9.18PVQIKK295 pKa = 10.19GGIPSIVLCNPGEE308 pKa = 4.25GSSYY312 pKa = 10.53IAFLNKK318 pKa = 9.95EE319 pKa = 3.85EE320 pKa = 4.09NAPLRR325 pKa = 11.84AWTQKK330 pKa = 9.91NAQFVILHH338 pKa = 6.13APLYY342 pKa = 10.56KK343 pKa = 9.81STAQDD348 pKa = 3.02SS349 pKa = 3.81
Molecular weight: 39.45 kDa
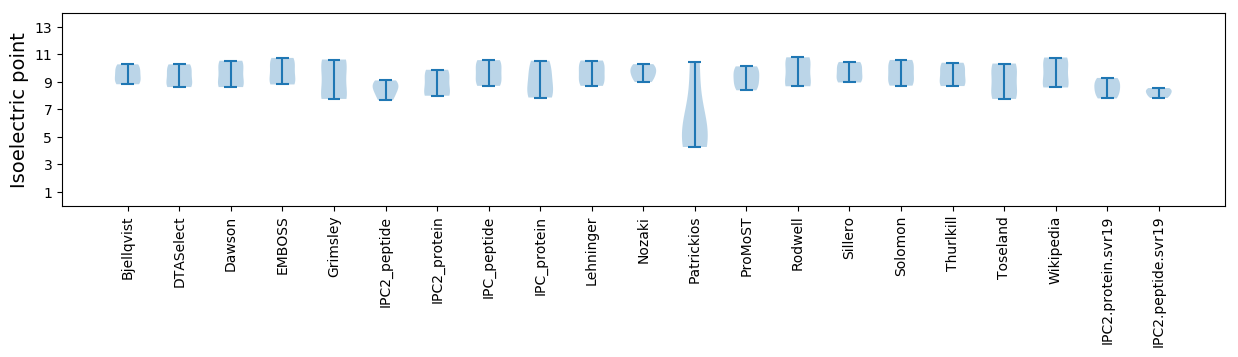
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9QJK8|Q9QJK8_9GEMI Capsid protein OS=Pepper golden mosaic virus-[CR] OX=223302 GN=AV1 PE=3 SV=1
MM1 pKa = 8.25DD2 pKa = 4.26SRR4 pKa = 11.84TGEE7 pKa = 4.39SITVRR12 pKa = 11.84QAEE15 pKa = 3.98NSVFIWKK22 pKa = 9.56VPNPLFFKK30 pKa = 9.82IQRR33 pKa = 11.84VEE35 pKa = 4.01APLDD39 pKa = 3.4TRR41 pKa = 11.84TRR43 pKa = 11.84IYY45 pKa = 10.5YY46 pKa = 9.57IEE48 pKa = 3.83VRR50 pKa = 11.84FNHH53 pKa = 5.71NLRR56 pKa = 11.84RR57 pKa = 11.84ALALHH62 pKa = 6.52KK63 pKa = 10.67SFLHH67 pKa = 4.9FQVWTTSMTASGTTYY82 pKa = 10.97LNRR85 pKa = 11.84FKK87 pKa = 10.85FRR89 pKa = 11.84IMLYY93 pKa = 8.49LHH95 pKa = 7.03RR96 pKa = 11.84LGAIGINNVIRR107 pKa = 11.84AVRR110 pKa = 11.84FATDD114 pKa = 3.02KK115 pKa = 11.32SYY117 pKa = 11.46VNYY120 pKa = 10.16VLEE123 pKa = 4.03HH124 pKa = 6.88HH125 pKa = 6.46EE126 pKa = 4.36IKK128 pKa = 10.88YY129 pKa = 10.66KK130 pKa = 10.9LYY132 pKa = 10.88
MM1 pKa = 8.25DD2 pKa = 4.26SRR4 pKa = 11.84TGEE7 pKa = 4.39SITVRR12 pKa = 11.84QAEE15 pKa = 3.98NSVFIWKK22 pKa = 9.56VPNPLFFKK30 pKa = 9.82IQRR33 pKa = 11.84VEE35 pKa = 4.01APLDD39 pKa = 3.4TRR41 pKa = 11.84TRR43 pKa = 11.84IYY45 pKa = 10.5YY46 pKa = 9.57IEE48 pKa = 3.83VRR50 pKa = 11.84FNHH53 pKa = 5.71NLRR56 pKa = 11.84RR57 pKa = 11.84ALALHH62 pKa = 6.52KK63 pKa = 10.67SFLHH67 pKa = 4.9FQVWTTSMTASGTTYY82 pKa = 10.97LNRR85 pKa = 11.84FKK87 pKa = 10.85FRR89 pKa = 11.84IMLYY93 pKa = 8.49LHH95 pKa = 7.03RR96 pKa = 11.84LGAIGINNVIRR107 pKa = 11.84AVRR110 pKa = 11.84FATDD114 pKa = 3.02KK115 pKa = 11.32SYY117 pKa = 11.46VNYY120 pKa = 10.16VLEE123 pKa = 4.03HH124 pKa = 6.88HH125 pKa = 6.46EE126 pKa = 4.36IKK128 pKa = 10.88YY129 pKa = 10.66KK130 pKa = 10.9LYY132 pKa = 10.88
Molecular weight: 15.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
861 |
129 |
349 |
215.3 |
24.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.156 ± 0.601 | 1.974 ± 0.449 |
5.226 ± 0.736 | 3.717 ± 0.377 |
4.878 ± 0.748 | 5.459 ± 0.557 |
3.833 ± 0.44 | 6.039 ± 0.893 |
6.62 ± 0.687 | 6.039 ± 0.888 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.207 ± 0.836 | 6.039 ± 0.331 |
5.807 ± 1.205 | 3.717 ± 0.56 |
7.201 ± 1.315 | 7.666 ± 1.028 |
4.762 ± 0.795 | 6.272 ± 1.096 |
1.626 ± 0.185 | 4.762 ± 0.901 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
