
Sewage-associated circular DNA virus-27
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.05
Get precalculated fractions of proteins
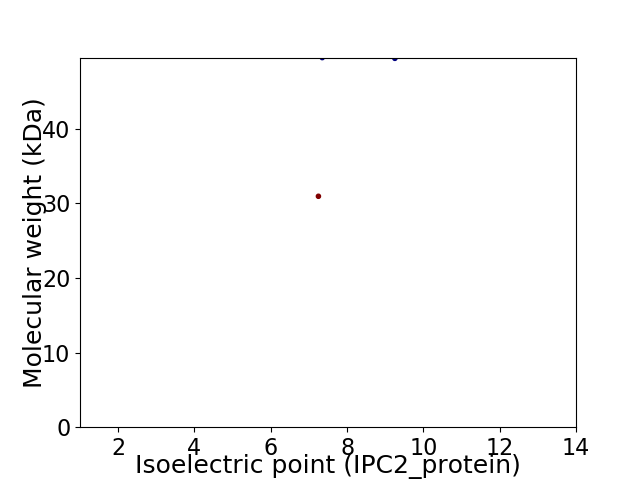
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UGZ0|A0A0B4UGZ0_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-27 OX=1592094 PE=4 SV=1
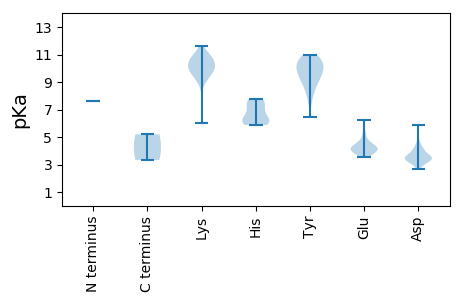
MM1 pKa = 7.63EE2 pKa = 5.26RR3 pKa = 11.84RR4 pKa = 11.84QGIYY8 pKa = 9.37WLLTIPFHH16 pKa = 6.43EE17 pKa = 4.47WTPYY21 pKa = 10.36LPPGVSWIRR30 pKa = 11.84GQLEE34 pKa = 4.14SGSEE38 pKa = 3.74SGYY41 pKa = 9.4LHH43 pKa = 5.88WQIVVALTTKK53 pKa = 10.49CSLRR57 pKa = 11.84GIKK60 pKa = 10.35GLFGDD65 pKa = 4.22STHH68 pKa = 7.05GEE70 pKa = 3.83LSRR73 pKa = 11.84TNRR76 pKa = 11.84ASVYY80 pKa = 7.78VWKK83 pKa = 10.64EE84 pKa = 3.56EE85 pKa = 3.81TRR87 pKa = 11.84IEE89 pKa = 4.15GTQFEE94 pKa = 4.62LGEE97 pKa = 4.23RR98 pKa = 11.84PFKK101 pKa = 10.85RR102 pKa = 11.84NCSKK106 pKa = 10.71DD107 pKa = 2.87WEE109 pKa = 4.66QIWEE113 pKa = 4.2CAQRR117 pKa = 11.84GDD119 pKa = 4.76LMSIPADD126 pKa = 3.44VRR128 pKa = 11.84VHH130 pKa = 6.39SYY132 pKa = 8.51RR133 pKa = 11.84TLRR136 pKa = 11.84AISADD141 pKa = 3.52YY142 pKa = 10.83SKK144 pKa = 10.17PLPMVRR150 pKa = 11.84DD151 pKa = 3.09IHH153 pKa = 7.77VYY155 pKa = 9.98IGPTATGKK163 pKa = 9.78SRR165 pKa = 11.84RR166 pKa = 11.84AWDD169 pKa = 3.51EE170 pKa = 3.81AGMEE174 pKa = 4.72AYY176 pKa = 10.5CKK178 pKa = 10.64DD179 pKa = 3.72PNSKK183 pKa = 9.69FWCGYY188 pKa = 9.01SGQPNIVLDD197 pKa = 4.37EE198 pKa = 3.89YY199 pKa = 10.81RR200 pKa = 11.84GRR202 pKa = 11.84IDD204 pKa = 3.65VSHH207 pKa = 7.42LLRR210 pKa = 11.84WFDD213 pKa = 3.73RR214 pKa = 11.84YY215 pKa = 9.57PVNVEE220 pKa = 3.73IKK222 pKa = 9.78GSSVPLCAEE231 pKa = 4.28SFWITTNLDD240 pKa = 2.7VDD242 pKa = 3.33QWYY245 pKa = 9.3PDD247 pKa = 3.58LDD249 pKa = 3.8ATTLEE254 pKa = 3.98ALRR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84LKK261 pKa = 9.11ITYY264 pKa = 8.88FRR266 pKa = 5.2
MM1 pKa = 7.63EE2 pKa = 5.26RR3 pKa = 11.84RR4 pKa = 11.84QGIYY8 pKa = 9.37WLLTIPFHH16 pKa = 6.43EE17 pKa = 4.47WTPYY21 pKa = 10.36LPPGVSWIRR30 pKa = 11.84GQLEE34 pKa = 4.14SGSEE38 pKa = 3.74SGYY41 pKa = 9.4LHH43 pKa = 5.88WQIVVALTTKK53 pKa = 10.49CSLRR57 pKa = 11.84GIKK60 pKa = 10.35GLFGDD65 pKa = 4.22STHH68 pKa = 7.05GEE70 pKa = 3.83LSRR73 pKa = 11.84TNRR76 pKa = 11.84ASVYY80 pKa = 7.78VWKK83 pKa = 10.64EE84 pKa = 3.56EE85 pKa = 3.81TRR87 pKa = 11.84IEE89 pKa = 4.15GTQFEE94 pKa = 4.62LGEE97 pKa = 4.23RR98 pKa = 11.84PFKK101 pKa = 10.85RR102 pKa = 11.84NCSKK106 pKa = 10.71DD107 pKa = 2.87WEE109 pKa = 4.66QIWEE113 pKa = 4.2CAQRR117 pKa = 11.84GDD119 pKa = 4.76LMSIPADD126 pKa = 3.44VRR128 pKa = 11.84VHH130 pKa = 6.39SYY132 pKa = 8.51RR133 pKa = 11.84TLRR136 pKa = 11.84AISADD141 pKa = 3.52YY142 pKa = 10.83SKK144 pKa = 10.17PLPMVRR150 pKa = 11.84DD151 pKa = 3.09IHH153 pKa = 7.77VYY155 pKa = 9.98IGPTATGKK163 pKa = 9.78SRR165 pKa = 11.84RR166 pKa = 11.84AWDD169 pKa = 3.51EE170 pKa = 3.81AGMEE174 pKa = 4.72AYY176 pKa = 10.5CKK178 pKa = 10.64DD179 pKa = 3.72PNSKK183 pKa = 9.69FWCGYY188 pKa = 9.01SGQPNIVLDD197 pKa = 4.37EE198 pKa = 3.89YY199 pKa = 10.81RR200 pKa = 11.84GRR202 pKa = 11.84IDD204 pKa = 3.65VSHH207 pKa = 7.42LLRR210 pKa = 11.84WFDD213 pKa = 3.73RR214 pKa = 11.84YY215 pKa = 9.57PVNVEE220 pKa = 3.73IKK222 pKa = 9.78GSSVPLCAEE231 pKa = 4.28SFWITTNLDD240 pKa = 2.7VDD242 pKa = 3.33QWYY245 pKa = 9.3PDD247 pKa = 3.58LDD249 pKa = 3.8ATTLEE254 pKa = 3.98ALRR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84LKK261 pKa = 9.11ITYY264 pKa = 8.88FRR266 pKa = 5.2
Molecular weight: 30.95 kDa
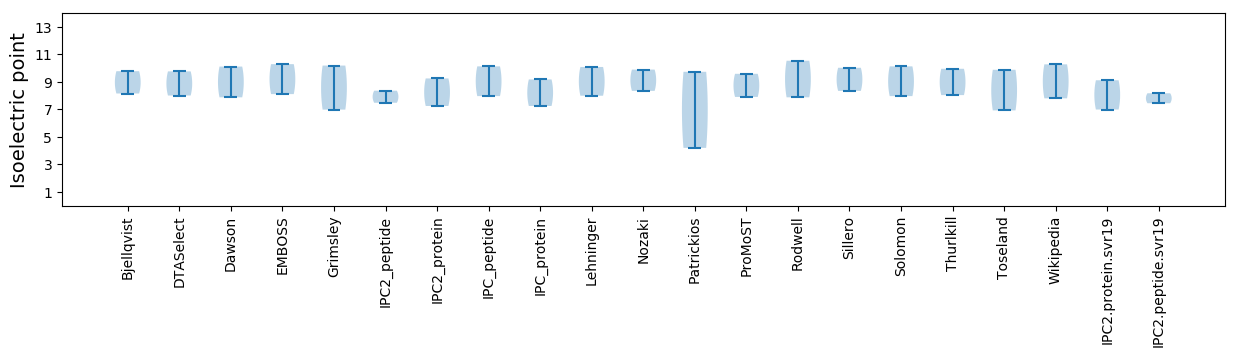
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UGZ0|A0A0B4UGZ0_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-27 OX=1592094 PE=4 SV=1
MM1 pKa = 7.63TIRR4 pKa = 11.84HH5 pKa = 5.88EE6 pKa = 4.16LAGLAGGTLGFIHH19 pKa = 7.74DD20 pKa = 4.14NTRR23 pKa = 11.84GAIKK27 pKa = 10.26GYY29 pKa = 10.32YY30 pKa = 9.38LGKK33 pKa = 10.02QLSKK37 pKa = 11.44SMAPIPKK44 pKa = 9.38KK45 pKa = 10.3RR46 pKa = 11.84RR47 pKa = 11.84NSYY50 pKa = 7.95STPSTRR56 pKa = 11.84GTITNGILSRR66 pKa = 11.84RR67 pKa = 11.84SSNPFDD73 pKa = 4.11RR74 pKa = 11.84RR75 pKa = 11.84LSDD78 pKa = 3.15VSMRR82 pKa = 11.84SAKK85 pKa = 8.64GTYY88 pKa = 9.41RR89 pKa = 11.84AFTGRR94 pKa = 11.84GTGSASASGYY104 pKa = 10.41RR105 pKa = 11.84SISGNPAAIKK115 pKa = 9.46KK116 pKa = 9.25RR117 pKa = 11.84KK118 pKa = 9.26RR119 pKa = 11.84KK120 pKa = 9.91GVVLKK125 pKa = 10.42KK126 pKa = 9.99KK127 pKa = 10.34RR128 pKa = 11.84RR129 pKa = 11.84VKK131 pKa = 10.74VSGKK135 pKa = 9.65FKK137 pKa = 10.7RR138 pKa = 11.84KK139 pKa = 5.99VHH141 pKa = 6.14KK142 pKa = 10.82ALDD145 pKa = 3.38EE146 pKa = 4.67KK147 pKa = 10.94IYY149 pKa = 10.46GKK151 pKa = 9.57HH152 pKa = 6.0LKK154 pKa = 9.07VHH156 pKa = 6.31YY157 pKa = 10.02DD158 pKa = 3.43RR159 pKa = 11.84LPATGSYY166 pKa = 8.48QQSIKK171 pKa = 11.08DD172 pKa = 3.74FGHH175 pKa = 7.21LFTPLRR181 pKa = 11.84MIAAADD187 pKa = 3.48VLFNKK192 pKa = 10.17APVVEE197 pKa = 6.26FPATTDD203 pKa = 3.32IKK205 pKa = 10.42WNNSFIRR212 pKa = 11.84QDD214 pKa = 3.47YY215 pKa = 10.04VINSWASFEE224 pKa = 5.26LKK226 pKa = 10.29NCSQRR231 pKa = 11.84TYY233 pKa = 8.77TIKK236 pKa = 10.04MYY238 pKa = 10.28EE239 pKa = 4.24CKK241 pKa = 9.95PLNKK245 pKa = 9.94AVLPVTNDD253 pKa = 3.47AYY255 pKa = 10.89ADD257 pKa = 4.19WIRR260 pKa = 11.84GMLAMQGLQTNPASNTPNTLYY281 pKa = 10.6SVPFDD286 pKa = 3.34SPQFNQCWKK295 pKa = 11.08AEE297 pKa = 4.06VTTCILEE304 pKa = 4.38PGQSNTFFVQGPNQQSIDD322 pKa = 3.35WQKK325 pKa = 10.86YY326 pKa = 6.44VYY328 pKa = 9.6EE329 pKa = 4.12NSGFVDD335 pKa = 3.23VMNEE339 pKa = 3.54FATWSRR345 pKa = 11.84DD346 pKa = 3.39VFFVYY351 pKa = 10.96YY352 pKa = 9.84EE353 pKa = 4.28DD354 pKa = 5.88LVTTDD359 pKa = 3.39GNNVGRR365 pKa = 11.84IVSGPVTGGGGVAIEE380 pKa = 4.1FKK382 pKa = 10.92EE383 pKa = 4.14FFKK386 pKa = 11.58LEE388 pKa = 4.15VPASAGFTYY397 pKa = 9.98PASTAAGVKK406 pKa = 9.48QQLDD410 pKa = 3.39NKK412 pKa = 9.84LPTKK416 pKa = 10.04IISVFPQIVVGNVVDD431 pKa = 3.98VAEE434 pKa = 4.5EE435 pKa = 4.07NPAVSTNPYY444 pKa = 10.12DD445 pKa = 3.33
MM1 pKa = 7.63TIRR4 pKa = 11.84HH5 pKa = 5.88EE6 pKa = 4.16LAGLAGGTLGFIHH19 pKa = 7.74DD20 pKa = 4.14NTRR23 pKa = 11.84GAIKK27 pKa = 10.26GYY29 pKa = 10.32YY30 pKa = 9.38LGKK33 pKa = 10.02QLSKK37 pKa = 11.44SMAPIPKK44 pKa = 9.38KK45 pKa = 10.3RR46 pKa = 11.84RR47 pKa = 11.84NSYY50 pKa = 7.95STPSTRR56 pKa = 11.84GTITNGILSRR66 pKa = 11.84RR67 pKa = 11.84SSNPFDD73 pKa = 4.11RR74 pKa = 11.84RR75 pKa = 11.84LSDD78 pKa = 3.15VSMRR82 pKa = 11.84SAKK85 pKa = 8.64GTYY88 pKa = 9.41RR89 pKa = 11.84AFTGRR94 pKa = 11.84GTGSASASGYY104 pKa = 10.41RR105 pKa = 11.84SISGNPAAIKK115 pKa = 9.46KK116 pKa = 9.25RR117 pKa = 11.84KK118 pKa = 9.26RR119 pKa = 11.84KK120 pKa = 9.91GVVLKK125 pKa = 10.42KK126 pKa = 9.99KK127 pKa = 10.34RR128 pKa = 11.84RR129 pKa = 11.84VKK131 pKa = 10.74VSGKK135 pKa = 9.65FKK137 pKa = 10.7RR138 pKa = 11.84KK139 pKa = 5.99VHH141 pKa = 6.14KK142 pKa = 10.82ALDD145 pKa = 3.38EE146 pKa = 4.67KK147 pKa = 10.94IYY149 pKa = 10.46GKK151 pKa = 9.57HH152 pKa = 6.0LKK154 pKa = 9.07VHH156 pKa = 6.31YY157 pKa = 10.02DD158 pKa = 3.43RR159 pKa = 11.84LPATGSYY166 pKa = 8.48QQSIKK171 pKa = 11.08DD172 pKa = 3.74FGHH175 pKa = 7.21LFTPLRR181 pKa = 11.84MIAAADD187 pKa = 3.48VLFNKK192 pKa = 10.17APVVEE197 pKa = 6.26FPATTDD203 pKa = 3.32IKK205 pKa = 10.42WNNSFIRR212 pKa = 11.84QDD214 pKa = 3.47YY215 pKa = 10.04VINSWASFEE224 pKa = 5.26LKK226 pKa = 10.29NCSQRR231 pKa = 11.84TYY233 pKa = 8.77TIKK236 pKa = 10.04MYY238 pKa = 10.28EE239 pKa = 4.24CKK241 pKa = 9.95PLNKK245 pKa = 9.94AVLPVTNDD253 pKa = 3.47AYY255 pKa = 10.89ADD257 pKa = 4.19WIRR260 pKa = 11.84GMLAMQGLQTNPASNTPNTLYY281 pKa = 10.6SVPFDD286 pKa = 3.34SPQFNQCWKK295 pKa = 11.08AEE297 pKa = 4.06VTTCILEE304 pKa = 4.38PGQSNTFFVQGPNQQSIDD322 pKa = 3.35WQKK325 pKa = 10.86YY326 pKa = 6.44VYY328 pKa = 9.6EE329 pKa = 4.12NSGFVDD335 pKa = 3.23VMNEE339 pKa = 3.54FATWSRR345 pKa = 11.84DD346 pKa = 3.39VFFVYY351 pKa = 10.96YY352 pKa = 9.84EE353 pKa = 4.28DD354 pKa = 5.88LVTTDD359 pKa = 3.39GNNVGRR365 pKa = 11.84IVSGPVTGGGGVAIEE380 pKa = 4.1FKK382 pKa = 10.92EE383 pKa = 4.14FFKK386 pKa = 11.58LEE388 pKa = 4.15VPASAGFTYY397 pKa = 9.98PASTAAGVKK406 pKa = 9.48QQLDD410 pKa = 3.39NKK412 pKa = 9.84LPTKK416 pKa = 10.04IISVFPQIVVGNVVDD431 pKa = 3.98VAEE434 pKa = 4.5EE435 pKa = 4.07NPAVSTNPYY444 pKa = 10.12DD445 pKa = 3.33
Molecular weight: 49.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
711 |
266 |
445 |
355.5 |
40.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.47 ± 0.854 | 1.406 ± 0.458 |
4.923 ± 0.386 | 4.641 ± 1.146 |
4.36 ± 0.73 | 7.595 ± 0.244 |
1.688 ± 0.306 | 5.485 ± 0.286 |
6.47 ± 1.259 | 6.61 ± 0.895 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.688 ± 0.099 | 4.501 ± 1.211 |
5.345 ± 0.044 | 3.516 ± 0.274 |
6.892 ± 1.149 | 7.736 ± 0.117 |
6.61 ± 0.321 | 7.032 ± 0.751 |
2.532 ± 1.068 | 4.501 ± 0.208 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
