
Dietzia sp. UCD-THP
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Dietziaceae; Dietzia; unclassified Dietzia
Average proteome isoelectric point is 5.83
Get precalculated fractions of proteins
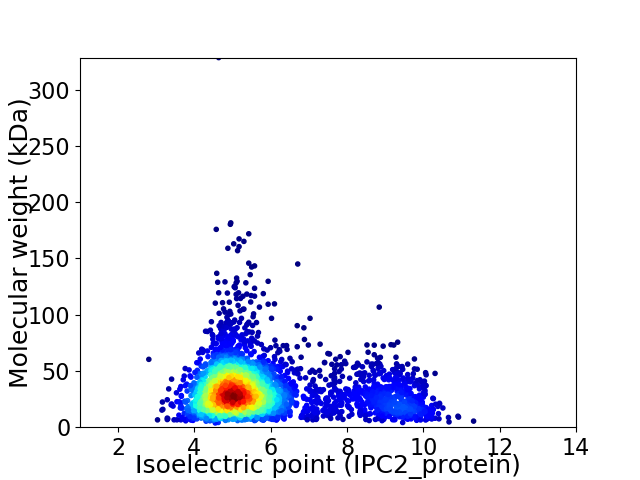
Virtual 2D-PAGE plot for 3489 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A022LPY7|A0A022LPY7_9ACTN Monooxygenase OS=Dietzia sp. UCD-THP OX=1292020 GN=H483_0100790 PE=4 SV=1
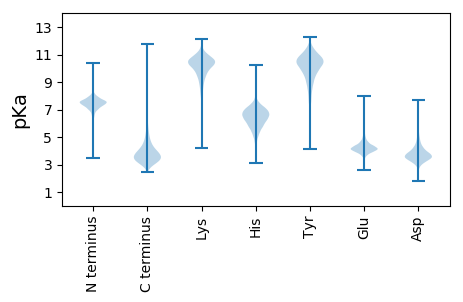
MM1 pKa = 7.37AVHH4 pKa = 6.85GRR6 pKa = 11.84FLKK9 pKa = 10.75AAAAASMVAMVSAACTNGGGGEE31 pKa = 4.09SDD33 pKa = 3.76EE34 pKa = 4.12QTEE37 pKa = 4.35AGTSGVGPITFAMGSNDD54 pKa = 3.55TDD56 pKa = 3.49KK57 pKa = 11.01LRR59 pKa = 11.84PVIDD63 pKa = 3.05QWNAEE68 pKa = 4.15NPDD71 pKa = 3.4QEE73 pKa = 4.4VTLRR77 pKa = 11.84EE78 pKa = 4.21LPAEE82 pKa = 3.95QDD84 pKa = 3.61GQRR87 pKa = 11.84DD88 pKa = 3.77TLTQSLQTEE97 pKa = 4.16SGEE100 pKa = 4.12YY101 pKa = 10.45DD102 pKa = 3.78VFALDD107 pKa = 3.9VTDD110 pKa = 3.61TAYY113 pKa = 10.56FAANGWLQPIEE124 pKa = 4.46GDD126 pKa = 3.9TEE128 pKa = 4.43IDD130 pKa = 3.14TSGLIEE136 pKa = 4.33AAVDD140 pKa = 3.4SATYY144 pKa = 10.68NGTLYY149 pKa = 10.96GIPQNTNAQLLYY161 pKa = 10.73YY162 pKa = 9.59RR163 pKa = 11.84TDD165 pKa = 3.74LQPEE169 pKa = 4.28APADD173 pKa = 3.52WDD175 pKa = 3.85ALVASCEE182 pKa = 4.05AAEE185 pKa = 4.65GADD188 pKa = 3.71VDD190 pKa = 4.64CLQTQLSLYY199 pKa = 10.29EE200 pKa = 4.32GLTVAATQFIHH211 pKa = 5.87SWGGRR216 pKa = 11.84VVGDD220 pKa = 4.2DD221 pKa = 3.65GQTPEE226 pKa = 5.55LDD228 pKa = 3.26TDD230 pKa = 3.49QARR233 pKa = 11.84AGLTALVDD241 pKa = 4.42AYY243 pKa = 11.01QNGVIPPQTDD253 pKa = 2.65AFTEE257 pKa = 4.26EE258 pKa = 3.88EE259 pKa = 4.28TAQGFLAGEE268 pKa = 4.18TMYY271 pKa = 10.87SYY273 pKa = 10.98NWPYY277 pKa = 10.06MYY279 pKa = 10.45EE280 pKa = 4.18SGQSDD285 pKa = 3.76PTSQVQGRR293 pKa = 11.84FEE295 pKa = 3.87VAPIVGQDD303 pKa = 3.17GAGRR307 pKa = 11.84STLGGYY313 pKa = 10.17NNAINVFSEE322 pKa = 4.26NKK324 pKa = 8.83ATAQAFLEE332 pKa = 4.47YY333 pKa = 9.91IISEE337 pKa = 4.17DD338 pKa = 3.48VQMSFAEE345 pKa = 4.23QSFPPVLSSIYY356 pKa = 10.34DD357 pKa = 3.68DD358 pKa = 4.62EE359 pKa = 6.06ALQQQFPYY367 pKa = 10.12MPALKK372 pKa = 10.22AALEE376 pKa = 4.16NAEE379 pKa = 4.36PRR381 pKa = 11.84PVTPYY386 pKa = 10.34YY387 pKa = 9.83PAVSKK392 pKa = 10.85AIQDD396 pKa = 3.48NTFAALRR403 pKa = 11.84GQKK406 pKa = 9.8NVEE409 pKa = 3.85QALTDD414 pKa = 3.36MSAAIDD420 pKa = 3.54QAAQQ424 pKa = 3.04
MM1 pKa = 7.37AVHH4 pKa = 6.85GRR6 pKa = 11.84FLKK9 pKa = 10.75AAAAASMVAMVSAACTNGGGGEE31 pKa = 4.09SDD33 pKa = 3.76EE34 pKa = 4.12QTEE37 pKa = 4.35AGTSGVGPITFAMGSNDD54 pKa = 3.55TDD56 pKa = 3.49KK57 pKa = 11.01LRR59 pKa = 11.84PVIDD63 pKa = 3.05QWNAEE68 pKa = 4.15NPDD71 pKa = 3.4QEE73 pKa = 4.4VTLRR77 pKa = 11.84EE78 pKa = 4.21LPAEE82 pKa = 3.95QDD84 pKa = 3.61GQRR87 pKa = 11.84DD88 pKa = 3.77TLTQSLQTEE97 pKa = 4.16SGEE100 pKa = 4.12YY101 pKa = 10.45DD102 pKa = 3.78VFALDD107 pKa = 3.9VTDD110 pKa = 3.61TAYY113 pKa = 10.56FAANGWLQPIEE124 pKa = 4.46GDD126 pKa = 3.9TEE128 pKa = 4.43IDD130 pKa = 3.14TSGLIEE136 pKa = 4.33AAVDD140 pKa = 3.4SATYY144 pKa = 10.68NGTLYY149 pKa = 10.96GIPQNTNAQLLYY161 pKa = 10.73YY162 pKa = 9.59RR163 pKa = 11.84TDD165 pKa = 3.74LQPEE169 pKa = 4.28APADD173 pKa = 3.52WDD175 pKa = 3.85ALVASCEE182 pKa = 4.05AAEE185 pKa = 4.65GADD188 pKa = 3.71VDD190 pKa = 4.64CLQTQLSLYY199 pKa = 10.29EE200 pKa = 4.32GLTVAATQFIHH211 pKa = 5.87SWGGRR216 pKa = 11.84VVGDD220 pKa = 4.2DD221 pKa = 3.65GQTPEE226 pKa = 5.55LDD228 pKa = 3.26TDD230 pKa = 3.49QARR233 pKa = 11.84AGLTALVDD241 pKa = 4.42AYY243 pKa = 11.01QNGVIPPQTDD253 pKa = 2.65AFTEE257 pKa = 4.26EE258 pKa = 3.88EE259 pKa = 4.28TAQGFLAGEE268 pKa = 4.18TMYY271 pKa = 10.87SYY273 pKa = 10.98NWPYY277 pKa = 10.06MYY279 pKa = 10.45EE280 pKa = 4.18SGQSDD285 pKa = 3.76PTSQVQGRR293 pKa = 11.84FEE295 pKa = 3.87VAPIVGQDD303 pKa = 3.17GAGRR307 pKa = 11.84STLGGYY313 pKa = 10.17NNAINVFSEE322 pKa = 4.26NKK324 pKa = 8.83ATAQAFLEE332 pKa = 4.47YY333 pKa = 9.91IISEE337 pKa = 4.17DD338 pKa = 3.48VQMSFAEE345 pKa = 4.23QSFPPVLSSIYY356 pKa = 10.34DD357 pKa = 3.68DD358 pKa = 4.62EE359 pKa = 6.06ALQQQFPYY367 pKa = 10.12MPALKK372 pKa = 10.22AALEE376 pKa = 4.16NAEE379 pKa = 4.36PRR381 pKa = 11.84PVTPYY386 pKa = 10.34YY387 pKa = 9.83PAVSKK392 pKa = 10.85AIQDD396 pKa = 3.48NTFAALRR403 pKa = 11.84GQKK406 pKa = 9.8NVEE409 pKa = 3.85QALTDD414 pKa = 3.36MSAAIDD420 pKa = 3.54QAAQQ424 pKa = 3.04
Molecular weight: 45.46 kDa
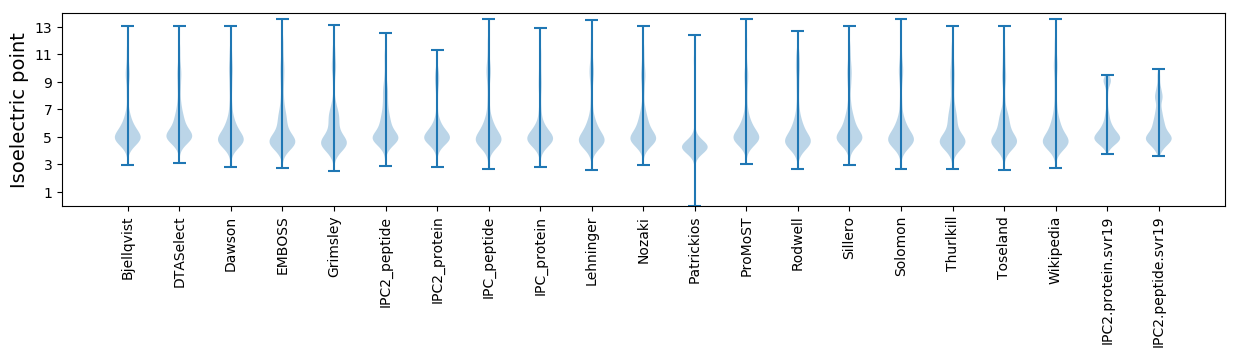
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A022LUP8|A0A022LUP8_9ACTN Acyl dehydratase OS=Dietzia sp. UCD-THP OX=1292020 GN=H483_0100935 PE=3 SV=1
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84AKK17 pKa = 9.45VHH19 pKa = 5.31GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84GIVGARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.68GRR42 pKa = 11.84ASLTAA47 pKa = 4.1
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84AKK17 pKa = 9.45VHH19 pKa = 5.31GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84GIVGARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.68GRR42 pKa = 11.84ASLTAA47 pKa = 4.1
Molecular weight: 5.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1118474 |
32 |
3134 |
320.6 |
34.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.843 ± 0.056 | 0.753 ± 0.011 |
6.614 ± 0.034 | 5.865 ± 0.035 |
2.757 ± 0.022 | 9.551 ± 0.045 |
2.196 ± 0.021 | 3.864 ± 0.029 |
1.733 ± 0.027 | 9.708 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.015 ± 0.015 | 1.728 ± 0.02 |
5.771 ± 0.034 | 2.562 ± 0.022 |
7.767 ± 0.047 | 5.623 ± 0.023 |
6.179 ± 0.027 | 9.125 ± 0.044 |
1.437 ± 0.017 | 1.907 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
