
Influenza B virus (strain B/Singapore/222/1979)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Insthoviricetes; Articulavirales; Orthomyxoviridae; Betainfluenzavirus; Influenza B virus
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
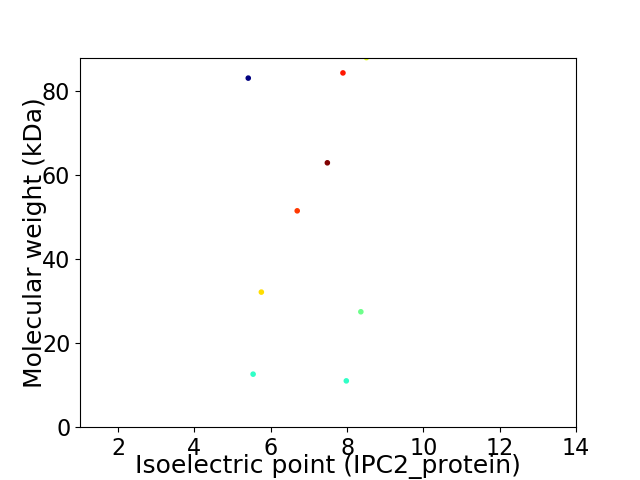
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
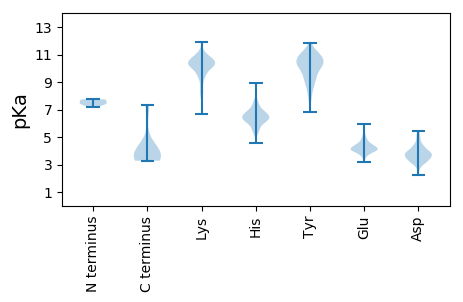
Protein with the lowest isoelectric point:
>sp|P12600|NS1_INBSI Non-structural protein 1 OS=Influenza B virus (strain B/Singapore/222/1979) OX=107417 GN=NS PE=3 SV=1
MM1 pKa = 7.72DD2 pKa = 4.03TFITRR7 pKa = 11.84NFQTTIIQKK16 pKa = 10.29AKK18 pKa = 8.58NTMAEE23 pKa = 4.07FMEE26 pKa = 5.03DD27 pKa = 3.9PEE29 pKa = 4.82LQPAMLFNICVHH41 pKa = 6.68LEE43 pKa = 3.78VCYY46 pKa = 10.78VISDD50 pKa = 3.68MNFLDD55 pKa = 4.43EE56 pKa = 4.43EE57 pKa = 4.77GKK59 pKa = 10.67AYY61 pKa = 8.15TALEE65 pKa = 4.33GQGKK69 pKa = 7.68EE70 pKa = 3.94QNLRR74 pKa = 11.84PQYY77 pKa = 10.54EE78 pKa = 4.57VIEE81 pKa = 4.19GMPRR85 pKa = 11.84NIAWMVQRR93 pKa = 11.84SLAQEE98 pKa = 4.19HH99 pKa = 6.59GIEE102 pKa = 4.28TPKK105 pKa = 10.97YY106 pKa = 9.38LADD109 pKa = 3.81LFDD112 pKa = 4.22YY113 pKa = 8.3KK114 pKa = 9.99TKK116 pKa = 10.82RR117 pKa = 11.84FIEE120 pKa = 4.0VGITKK125 pKa = 10.32GLADD129 pKa = 4.42DD130 pKa = 4.63YY131 pKa = 11.19FWKK134 pKa = 10.59KK135 pKa = 10.28KK136 pKa = 9.87EE137 pKa = 3.82KK138 pKa = 10.49LGNSMEE144 pKa = 4.41LMIFSYY150 pKa = 10.74NQDD153 pKa = 2.93YY154 pKa = 11.2SLSNEE159 pKa = 3.9SSLDD163 pKa = 3.56EE164 pKa = 4.4EE165 pKa = 4.86GKK167 pKa = 10.97GRR169 pKa = 11.84VLSRR173 pKa = 11.84LTEE176 pKa = 4.09LQAEE180 pKa = 4.6LSLKK184 pKa = 10.43NLWQVLIGEE193 pKa = 4.28EE194 pKa = 4.54DD195 pKa = 3.28IEE197 pKa = 4.69KK198 pKa = 11.15GIDD201 pKa = 3.45FKK203 pKa = 11.36LGQTISRR210 pKa = 11.84LRR212 pKa = 11.84DD213 pKa = 3.1ISVPAGFSNFEE224 pKa = 4.0GMRR227 pKa = 11.84SYY229 pKa = 11.04IDD231 pKa = 4.25NIDD234 pKa = 3.38PKK236 pKa = 11.14GAIEE240 pKa = 4.13RR241 pKa = 11.84NLARR245 pKa = 11.84MSPLVSVTPKK255 pKa = 9.85KK256 pKa = 10.83LKK258 pKa = 10.23WEE260 pKa = 4.3DD261 pKa = 3.41LRR263 pKa = 11.84PIGPHH268 pKa = 6.43IYY270 pKa = 9.79NHH272 pKa = 6.28EE273 pKa = 4.17LPEE276 pKa = 4.04VPYY279 pKa = 10.96NAFLLMSDD287 pKa = 3.99EE288 pKa = 4.95LGLANMTEE296 pKa = 4.57GKK298 pKa = 9.69SKK300 pKa = 10.66KK301 pKa = 9.72PKK303 pKa = 9.48TLAKK307 pKa = 10.0EE308 pKa = 4.07CLEE311 pKa = 4.39KK312 pKa = 11.23YY313 pKa = 8.63STLRR317 pKa = 11.84DD318 pKa = 3.36QTDD321 pKa = 4.33PILIMKK327 pKa = 9.41SEE329 pKa = 4.03KK330 pKa = 10.78ANEE333 pKa = 3.8NFLWKK338 pKa = 10.42LWRR341 pKa = 11.84DD342 pKa = 3.76CVNTISNEE350 pKa = 3.98EE351 pKa = 4.1TSNEE355 pKa = 3.91LQKK358 pKa = 10.16TNYY361 pKa = 9.82AKK363 pKa = 10.3WATGDD368 pKa = 3.24GLTYY372 pKa = 10.2QKK374 pKa = 10.63IMKK377 pKa = 9.33EE378 pKa = 3.96VAIDD382 pKa = 3.9DD383 pKa = 3.68EE384 pKa = 5.15TMCQEE389 pKa = 4.25EE390 pKa = 4.1PKK392 pKa = 10.31IPNKK396 pKa = 10.07CRR398 pKa = 11.84VAAWVQTEE406 pKa = 4.14MNLLSTLTSKK416 pKa = 10.85RR417 pKa = 11.84ALDD420 pKa = 3.91LPEE423 pKa = 5.27IGPDD427 pKa = 3.43VAPVEE432 pKa = 4.56HH433 pKa = 6.53VGSEE437 pKa = 3.58RR438 pKa = 11.84RR439 pKa = 11.84KK440 pKa = 9.81YY441 pKa = 9.86FVNEE445 pKa = 3.34INYY448 pKa = 9.25CKK450 pKa = 10.56ASTVMMKK457 pKa = 10.46YY458 pKa = 10.74VLFHH462 pKa = 6.42TSLLNEE468 pKa = 4.68SNASMGKK475 pKa = 9.99YY476 pKa = 9.62KK477 pKa = 10.59VIPITNRR484 pKa = 11.84VNEE487 pKa = 4.2KK488 pKa = 10.96GEE490 pKa = 4.3SFDD493 pKa = 3.5MLYY496 pKa = 11.24GLAVKK501 pKa = 9.91GQSHH505 pKa = 6.71LRR507 pKa = 11.84GDD509 pKa = 3.87TDD511 pKa = 3.58VVTVVTFEE519 pKa = 4.42FSSTDD524 pKa = 3.3PRR526 pKa = 11.84VDD528 pKa = 3.05SGKK531 pKa = 8.47WPKK534 pKa = 9.4YY535 pKa = 6.84TVFRR539 pKa = 11.84IGSLFVSGRR548 pKa = 11.84EE549 pKa = 3.75KK550 pKa = 10.61SVYY553 pKa = 10.09LYY555 pKa = 11.01CRR557 pKa = 11.84VNGTNKK563 pKa = 9.61IQMKK567 pKa = 8.84WGMEE571 pKa = 3.68ARR573 pKa = 11.84RR574 pKa = 11.84CLLQSMQQMEE584 pKa = 5.02AIVEE588 pKa = 4.22QEE590 pKa = 4.1SSIQGYY596 pKa = 11.18DD597 pKa = 3.05MTKK600 pKa = 10.49ACFKK604 pKa = 9.93EE605 pKa = 4.19DD606 pKa = 3.17RR607 pKa = 11.84VNSPKK612 pKa = 10.09TFSIGTQEE620 pKa = 3.93GKK622 pKa = 10.13LVKK625 pKa = 10.6GSFGKK630 pKa = 10.38ALRR633 pKa = 11.84VIFTKK638 pKa = 10.82CLMHH642 pKa = 6.24YY643 pKa = 10.55VFGNAQLEE651 pKa = 4.61GFSAEE656 pKa = 4.01SRR658 pKa = 11.84RR659 pKa = 11.84LLLLIQALKK668 pKa = 10.37DD669 pKa = 3.68RR670 pKa = 11.84KK671 pKa = 10.3GPWVFDD677 pKa = 3.85LEE679 pKa = 4.38GMYY682 pKa = 10.41SGIEE686 pKa = 3.85EE687 pKa = 4.9CISNNPWVIQSAYY700 pKa = 8.73WFNEE704 pKa = 3.19WLGFEE709 pKa = 4.34KK710 pKa = 10.6EE711 pKa = 4.12GSKK714 pKa = 10.62VLEE717 pKa = 4.4SVDD720 pKa = 4.31EE721 pKa = 4.55IMDD724 pKa = 3.72EE725 pKa = 4.12
MM1 pKa = 7.72DD2 pKa = 4.03TFITRR7 pKa = 11.84NFQTTIIQKK16 pKa = 10.29AKK18 pKa = 8.58NTMAEE23 pKa = 4.07FMEE26 pKa = 5.03DD27 pKa = 3.9PEE29 pKa = 4.82LQPAMLFNICVHH41 pKa = 6.68LEE43 pKa = 3.78VCYY46 pKa = 10.78VISDD50 pKa = 3.68MNFLDD55 pKa = 4.43EE56 pKa = 4.43EE57 pKa = 4.77GKK59 pKa = 10.67AYY61 pKa = 8.15TALEE65 pKa = 4.33GQGKK69 pKa = 7.68EE70 pKa = 3.94QNLRR74 pKa = 11.84PQYY77 pKa = 10.54EE78 pKa = 4.57VIEE81 pKa = 4.19GMPRR85 pKa = 11.84NIAWMVQRR93 pKa = 11.84SLAQEE98 pKa = 4.19HH99 pKa = 6.59GIEE102 pKa = 4.28TPKK105 pKa = 10.97YY106 pKa = 9.38LADD109 pKa = 3.81LFDD112 pKa = 4.22YY113 pKa = 8.3KK114 pKa = 9.99TKK116 pKa = 10.82RR117 pKa = 11.84FIEE120 pKa = 4.0VGITKK125 pKa = 10.32GLADD129 pKa = 4.42DD130 pKa = 4.63YY131 pKa = 11.19FWKK134 pKa = 10.59KK135 pKa = 10.28KK136 pKa = 9.87EE137 pKa = 3.82KK138 pKa = 10.49LGNSMEE144 pKa = 4.41LMIFSYY150 pKa = 10.74NQDD153 pKa = 2.93YY154 pKa = 11.2SLSNEE159 pKa = 3.9SSLDD163 pKa = 3.56EE164 pKa = 4.4EE165 pKa = 4.86GKK167 pKa = 10.97GRR169 pKa = 11.84VLSRR173 pKa = 11.84LTEE176 pKa = 4.09LQAEE180 pKa = 4.6LSLKK184 pKa = 10.43NLWQVLIGEE193 pKa = 4.28EE194 pKa = 4.54DD195 pKa = 3.28IEE197 pKa = 4.69KK198 pKa = 11.15GIDD201 pKa = 3.45FKK203 pKa = 11.36LGQTISRR210 pKa = 11.84LRR212 pKa = 11.84DD213 pKa = 3.1ISVPAGFSNFEE224 pKa = 4.0GMRR227 pKa = 11.84SYY229 pKa = 11.04IDD231 pKa = 4.25NIDD234 pKa = 3.38PKK236 pKa = 11.14GAIEE240 pKa = 4.13RR241 pKa = 11.84NLARR245 pKa = 11.84MSPLVSVTPKK255 pKa = 9.85KK256 pKa = 10.83LKK258 pKa = 10.23WEE260 pKa = 4.3DD261 pKa = 3.41LRR263 pKa = 11.84PIGPHH268 pKa = 6.43IYY270 pKa = 9.79NHH272 pKa = 6.28EE273 pKa = 4.17LPEE276 pKa = 4.04VPYY279 pKa = 10.96NAFLLMSDD287 pKa = 3.99EE288 pKa = 4.95LGLANMTEE296 pKa = 4.57GKK298 pKa = 9.69SKK300 pKa = 10.66KK301 pKa = 9.72PKK303 pKa = 9.48TLAKK307 pKa = 10.0EE308 pKa = 4.07CLEE311 pKa = 4.39KK312 pKa = 11.23YY313 pKa = 8.63STLRR317 pKa = 11.84DD318 pKa = 3.36QTDD321 pKa = 4.33PILIMKK327 pKa = 9.41SEE329 pKa = 4.03KK330 pKa = 10.78ANEE333 pKa = 3.8NFLWKK338 pKa = 10.42LWRR341 pKa = 11.84DD342 pKa = 3.76CVNTISNEE350 pKa = 3.98EE351 pKa = 4.1TSNEE355 pKa = 3.91LQKK358 pKa = 10.16TNYY361 pKa = 9.82AKK363 pKa = 10.3WATGDD368 pKa = 3.24GLTYY372 pKa = 10.2QKK374 pKa = 10.63IMKK377 pKa = 9.33EE378 pKa = 3.96VAIDD382 pKa = 3.9DD383 pKa = 3.68EE384 pKa = 5.15TMCQEE389 pKa = 4.25EE390 pKa = 4.1PKK392 pKa = 10.31IPNKK396 pKa = 10.07CRR398 pKa = 11.84VAAWVQTEE406 pKa = 4.14MNLLSTLTSKK416 pKa = 10.85RR417 pKa = 11.84ALDD420 pKa = 3.91LPEE423 pKa = 5.27IGPDD427 pKa = 3.43VAPVEE432 pKa = 4.56HH433 pKa = 6.53VGSEE437 pKa = 3.58RR438 pKa = 11.84RR439 pKa = 11.84KK440 pKa = 9.81YY441 pKa = 9.86FVNEE445 pKa = 3.34INYY448 pKa = 9.25CKK450 pKa = 10.56ASTVMMKK457 pKa = 10.46YY458 pKa = 10.74VLFHH462 pKa = 6.42TSLLNEE468 pKa = 4.68SNASMGKK475 pKa = 9.99YY476 pKa = 9.62KK477 pKa = 10.59VIPITNRR484 pKa = 11.84VNEE487 pKa = 4.2KK488 pKa = 10.96GEE490 pKa = 4.3SFDD493 pKa = 3.5MLYY496 pKa = 11.24GLAVKK501 pKa = 9.91GQSHH505 pKa = 6.71LRR507 pKa = 11.84GDD509 pKa = 3.87TDD511 pKa = 3.58VVTVVTFEE519 pKa = 4.42FSSTDD524 pKa = 3.3PRR526 pKa = 11.84VDD528 pKa = 3.05SGKK531 pKa = 8.47WPKK534 pKa = 9.4YY535 pKa = 6.84TVFRR539 pKa = 11.84IGSLFVSGRR548 pKa = 11.84EE549 pKa = 3.75KK550 pKa = 10.61SVYY553 pKa = 10.09LYY555 pKa = 11.01CRR557 pKa = 11.84VNGTNKK563 pKa = 9.61IQMKK567 pKa = 8.84WGMEE571 pKa = 3.68ARR573 pKa = 11.84RR574 pKa = 11.84CLLQSMQQMEE584 pKa = 5.02AIVEE588 pKa = 4.22QEE590 pKa = 4.1SSIQGYY596 pKa = 11.18DD597 pKa = 3.05MTKK600 pKa = 10.49ACFKK604 pKa = 9.93EE605 pKa = 4.19DD606 pKa = 3.17RR607 pKa = 11.84VNSPKK612 pKa = 10.09TFSIGTQEE620 pKa = 3.93GKK622 pKa = 10.13LVKK625 pKa = 10.6GSFGKK630 pKa = 10.38ALRR633 pKa = 11.84VIFTKK638 pKa = 10.82CLMHH642 pKa = 6.24YY643 pKa = 10.55VFGNAQLEE651 pKa = 4.61GFSAEE656 pKa = 4.01SRR658 pKa = 11.84RR659 pKa = 11.84LLLLIQALKK668 pKa = 10.37DD669 pKa = 3.68RR670 pKa = 11.84KK671 pKa = 10.3GPWVFDD677 pKa = 3.85LEE679 pKa = 4.38GMYY682 pKa = 10.41SGIEE686 pKa = 3.85EE687 pKa = 4.9CISNNPWVIQSAYY700 pKa = 8.73WFNEE704 pKa = 3.19WLGFEE709 pKa = 4.34KK710 pKa = 10.6EE711 pKa = 4.12GSKK714 pKa = 10.62VLEE717 pKa = 4.4SVDD720 pKa = 4.31EE721 pKa = 4.55IMDD724 pKa = 3.72EE725 pKa = 4.12
Molecular weight: 83.09 kDa
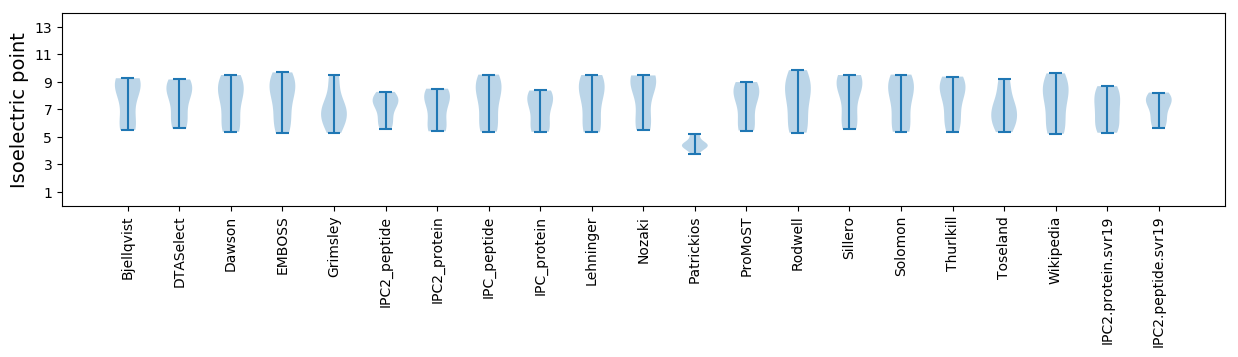
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9QLL3|Q9QLL3_INBSI Polymerase basic protein 2 OS=Influenza B virus (strain B/Singapore/222/1979) OX=107417 GN=PB2 PE=3 SV=1
MM1 pKa = 7.2TLAKK5 pKa = 10.05IEE7 pKa = 4.27LLKK10 pKa = 10.82QLLRR14 pKa = 11.84DD15 pKa = 3.91NEE17 pKa = 4.23AKK19 pKa = 9.7TVLKK23 pKa = 9.15QTTVDD28 pKa = 3.07QYY30 pKa = 11.86NIIRR34 pKa = 11.84KK35 pKa = 8.85FNTSRR40 pKa = 11.84IEE42 pKa = 4.0KK43 pKa = 10.24NPSLRR48 pKa = 11.84MKK50 pKa = 9.92WAMCSNFPLALTKK63 pKa = 10.87GDD65 pKa = 3.51MANRR69 pKa = 11.84IPLEE73 pKa = 3.92YY74 pKa = 10.33KK75 pKa = 10.63GIQLKK80 pKa = 9.38TNAEE84 pKa = 4.42DD85 pKa = 3.46IGTKK89 pKa = 10.43GQMCSIAAVTWWNTYY104 pKa = 10.1GPIGDD109 pKa = 3.74TEE111 pKa = 4.35GFEE114 pKa = 4.22KK115 pKa = 10.73VYY117 pKa = 11.02EE118 pKa = 4.3SFFLRR123 pKa = 11.84KK124 pKa = 8.78MRR126 pKa = 11.84LDD128 pKa = 3.16NATWGRR134 pKa = 11.84ITFGPVEE141 pKa = 4.17RR142 pKa = 11.84VRR144 pKa = 11.84KK145 pKa = 9.27RR146 pKa = 11.84VLLNPLTKK154 pKa = 10.41EE155 pKa = 3.99MPPDD159 pKa = 3.45EE160 pKa = 4.69ASNVIMEE167 pKa = 4.24ILFPKK172 pKa = 9.72EE173 pKa = 3.27AGIPRR178 pKa = 11.84EE179 pKa = 4.2STWIHH184 pKa = 6.43RR185 pKa = 11.84EE186 pKa = 3.8LIKK189 pKa = 10.54EE190 pKa = 3.96KK191 pKa = 10.54RR192 pKa = 11.84EE193 pKa = 3.94KK194 pKa = 11.07LKK196 pKa = 10.21GTMITPIVLAYY207 pKa = 8.9MLEE210 pKa = 4.26RR211 pKa = 11.84EE212 pKa = 4.28LVARR216 pKa = 11.84RR217 pKa = 11.84RR218 pKa = 11.84FLPVAGATSAEE229 pKa = 4.51FIEE232 pKa = 4.74MLHH235 pKa = 6.25CLQGEE240 pKa = 4.23NWRR243 pKa = 11.84QIYY246 pKa = 10.2HH247 pKa = 6.78PGGNKK252 pKa = 8.44LTEE255 pKa = 4.17SRR257 pKa = 11.84SQSMIVACRR266 pKa = 11.84KK267 pKa = 9.17IIRR270 pKa = 11.84RR271 pKa = 11.84SIVASNPLEE280 pKa = 4.1LAVEE284 pKa = 4.58IANKK288 pKa = 8.86TVIDD292 pKa = 3.95TEE294 pKa = 4.23PLKK297 pKa = 11.19SCLAAIDD304 pKa = 4.49GGDD307 pKa = 3.66VACDD311 pKa = 3.12IMRR314 pKa = 11.84AALGLKK320 pKa = 9.13IRR322 pKa = 11.84QRR324 pKa = 11.84QRR326 pKa = 11.84FGRR329 pKa = 11.84LEE331 pKa = 4.07LKK333 pKa = 10.28RR334 pKa = 11.84ISGRR338 pKa = 11.84GFKK341 pKa = 10.29NDD343 pKa = 3.3EE344 pKa = 4.59EE345 pKa = 4.2ILIGNGTIQKK355 pKa = 10.0IGIWDD360 pKa = 3.73GEE362 pKa = 4.47EE363 pKa = 3.99EE364 pKa = 4.11FHH366 pKa = 6.94VRR368 pKa = 11.84CGEE371 pKa = 3.88CRR373 pKa = 11.84GILKK377 pKa = 9.91KK378 pKa = 10.83SKK380 pKa = 9.48MRR382 pKa = 11.84MEE384 pKa = 4.32KK385 pKa = 10.82LLINSAKK392 pKa = 10.33KK393 pKa = 10.58EE394 pKa = 3.99DD395 pKa = 3.88MKK397 pKa = 11.48DD398 pKa = 3.71LIILCMVFSQDD409 pKa = 2.31TRR411 pKa = 11.84MFQGVRR417 pKa = 11.84GEE419 pKa = 4.12INFLNRR425 pKa = 11.84AGQLLSPMYY434 pKa = 10.01QLQRR438 pKa = 11.84YY439 pKa = 7.37FLNRR443 pKa = 11.84SNDD446 pKa = 3.9LFDD449 pKa = 2.96QWGYY453 pKa = 10.98EE454 pKa = 4.08EE455 pKa = 4.74SPKK458 pKa = 10.73ASEE461 pKa = 3.57LHH463 pKa = 6.88GINEE467 pKa = 4.33LMNASDD473 pKa = 3.92YY474 pKa = 8.88TLKK477 pKa = 11.05GVVVTKK483 pKa = 10.76NVIDD487 pKa = 4.64DD488 pKa = 4.65FSSTEE493 pKa = 4.04TEE495 pKa = 4.03KK496 pKa = 11.31VSITKK501 pKa = 10.01NLSLIKK507 pKa = 9.88RR508 pKa = 11.84TGEE511 pKa = 4.12VIMGANDD518 pKa = 3.09ISEE521 pKa = 5.01LEE523 pKa = 4.23SQAQLMITYY532 pKa = 7.68DD533 pKa = 3.87TPKK536 pKa = 8.17MWEE539 pKa = 3.77MGTTKK544 pKa = 10.43EE545 pKa = 4.29LVQNTYY551 pKa = 8.21QWVLKK556 pKa = 10.91NLVTLKK562 pKa = 10.93AQFLLGKK569 pKa = 9.96EE570 pKa = 5.88DD571 pKa = 3.54MFQWDD576 pKa = 3.33AFEE579 pKa = 4.66AFEE582 pKa = 5.18SIIPQKK588 pKa = 10.32MAGQYY593 pKa = 10.54SGFARR598 pKa = 11.84AVLKK602 pKa = 9.91QMRR605 pKa = 11.84DD606 pKa = 3.47QEE608 pKa = 4.51VMKK611 pKa = 10.1TDD613 pKa = 3.47QFIKK617 pKa = 10.67LLPFCFSPPKK627 pKa = 10.06LRR629 pKa = 11.84SNGEE633 pKa = 3.67PYY635 pKa = 10.49QFLRR639 pKa = 11.84LILKK643 pKa = 10.43GGGEE647 pKa = 4.11NFIEE651 pKa = 4.26VRR653 pKa = 11.84KK654 pKa = 10.13GSPLFSYY661 pKa = 10.68NPQTEE666 pKa = 4.12VLTICGRR673 pKa = 11.84MMSLKK678 pKa = 10.66GKK680 pKa = 10.2IEE682 pKa = 4.01DD683 pKa = 3.72EE684 pKa = 3.95EE685 pKa = 5.12RR686 pKa = 11.84NRR688 pKa = 11.84SMGNAVLAGFLVSGKK703 pKa = 10.1YY704 pKa = 10.53DD705 pKa = 3.9PDD707 pKa = 5.06LGDD710 pKa = 3.6FKK712 pKa = 10.87TIEE715 pKa = 4.29EE716 pKa = 4.4LEE718 pKa = 4.1KK719 pKa = 11.0LKK721 pKa = 10.75PGEE724 pKa = 3.97KK725 pKa = 10.57ANILLYY731 pKa = 10.07QGKK734 pKa = 7.97PVKK737 pKa = 9.66VVKK740 pKa = 10.37RR741 pKa = 11.84KK742 pKa = 9.92RR743 pKa = 11.84YY744 pKa = 8.87SALSNDD750 pKa = 2.67ISQGIKK756 pKa = 8.29RR757 pKa = 11.84QRR759 pKa = 11.84MTVEE763 pKa = 3.77SMGWALSS770 pKa = 3.61
MM1 pKa = 7.2TLAKK5 pKa = 10.05IEE7 pKa = 4.27LLKK10 pKa = 10.82QLLRR14 pKa = 11.84DD15 pKa = 3.91NEE17 pKa = 4.23AKK19 pKa = 9.7TVLKK23 pKa = 9.15QTTVDD28 pKa = 3.07QYY30 pKa = 11.86NIIRR34 pKa = 11.84KK35 pKa = 8.85FNTSRR40 pKa = 11.84IEE42 pKa = 4.0KK43 pKa = 10.24NPSLRR48 pKa = 11.84MKK50 pKa = 9.92WAMCSNFPLALTKK63 pKa = 10.87GDD65 pKa = 3.51MANRR69 pKa = 11.84IPLEE73 pKa = 3.92YY74 pKa = 10.33KK75 pKa = 10.63GIQLKK80 pKa = 9.38TNAEE84 pKa = 4.42DD85 pKa = 3.46IGTKK89 pKa = 10.43GQMCSIAAVTWWNTYY104 pKa = 10.1GPIGDD109 pKa = 3.74TEE111 pKa = 4.35GFEE114 pKa = 4.22KK115 pKa = 10.73VYY117 pKa = 11.02EE118 pKa = 4.3SFFLRR123 pKa = 11.84KK124 pKa = 8.78MRR126 pKa = 11.84LDD128 pKa = 3.16NATWGRR134 pKa = 11.84ITFGPVEE141 pKa = 4.17RR142 pKa = 11.84VRR144 pKa = 11.84KK145 pKa = 9.27RR146 pKa = 11.84VLLNPLTKK154 pKa = 10.41EE155 pKa = 3.99MPPDD159 pKa = 3.45EE160 pKa = 4.69ASNVIMEE167 pKa = 4.24ILFPKK172 pKa = 9.72EE173 pKa = 3.27AGIPRR178 pKa = 11.84EE179 pKa = 4.2STWIHH184 pKa = 6.43RR185 pKa = 11.84EE186 pKa = 3.8LIKK189 pKa = 10.54EE190 pKa = 3.96KK191 pKa = 10.54RR192 pKa = 11.84EE193 pKa = 3.94KK194 pKa = 11.07LKK196 pKa = 10.21GTMITPIVLAYY207 pKa = 8.9MLEE210 pKa = 4.26RR211 pKa = 11.84EE212 pKa = 4.28LVARR216 pKa = 11.84RR217 pKa = 11.84RR218 pKa = 11.84FLPVAGATSAEE229 pKa = 4.51FIEE232 pKa = 4.74MLHH235 pKa = 6.25CLQGEE240 pKa = 4.23NWRR243 pKa = 11.84QIYY246 pKa = 10.2HH247 pKa = 6.78PGGNKK252 pKa = 8.44LTEE255 pKa = 4.17SRR257 pKa = 11.84SQSMIVACRR266 pKa = 11.84KK267 pKa = 9.17IIRR270 pKa = 11.84RR271 pKa = 11.84SIVASNPLEE280 pKa = 4.1LAVEE284 pKa = 4.58IANKK288 pKa = 8.86TVIDD292 pKa = 3.95TEE294 pKa = 4.23PLKK297 pKa = 11.19SCLAAIDD304 pKa = 4.49GGDD307 pKa = 3.66VACDD311 pKa = 3.12IMRR314 pKa = 11.84AALGLKK320 pKa = 9.13IRR322 pKa = 11.84QRR324 pKa = 11.84QRR326 pKa = 11.84FGRR329 pKa = 11.84LEE331 pKa = 4.07LKK333 pKa = 10.28RR334 pKa = 11.84ISGRR338 pKa = 11.84GFKK341 pKa = 10.29NDD343 pKa = 3.3EE344 pKa = 4.59EE345 pKa = 4.2ILIGNGTIQKK355 pKa = 10.0IGIWDD360 pKa = 3.73GEE362 pKa = 4.47EE363 pKa = 3.99EE364 pKa = 4.11FHH366 pKa = 6.94VRR368 pKa = 11.84CGEE371 pKa = 3.88CRR373 pKa = 11.84GILKK377 pKa = 9.91KK378 pKa = 10.83SKK380 pKa = 9.48MRR382 pKa = 11.84MEE384 pKa = 4.32KK385 pKa = 10.82LLINSAKK392 pKa = 10.33KK393 pKa = 10.58EE394 pKa = 3.99DD395 pKa = 3.88MKK397 pKa = 11.48DD398 pKa = 3.71LIILCMVFSQDD409 pKa = 2.31TRR411 pKa = 11.84MFQGVRR417 pKa = 11.84GEE419 pKa = 4.12INFLNRR425 pKa = 11.84AGQLLSPMYY434 pKa = 10.01QLQRR438 pKa = 11.84YY439 pKa = 7.37FLNRR443 pKa = 11.84SNDD446 pKa = 3.9LFDD449 pKa = 2.96QWGYY453 pKa = 10.98EE454 pKa = 4.08EE455 pKa = 4.74SPKK458 pKa = 10.73ASEE461 pKa = 3.57LHH463 pKa = 6.88GINEE467 pKa = 4.33LMNASDD473 pKa = 3.92YY474 pKa = 8.88TLKK477 pKa = 11.05GVVVTKK483 pKa = 10.76NVIDD487 pKa = 4.64DD488 pKa = 4.65FSSTEE493 pKa = 4.04TEE495 pKa = 4.03KK496 pKa = 11.31VSITKK501 pKa = 10.01NLSLIKK507 pKa = 9.88RR508 pKa = 11.84TGEE511 pKa = 4.12VIMGANDD518 pKa = 3.09ISEE521 pKa = 5.01LEE523 pKa = 4.23SQAQLMITYY532 pKa = 7.68DD533 pKa = 3.87TPKK536 pKa = 8.17MWEE539 pKa = 3.77MGTTKK544 pKa = 10.43EE545 pKa = 4.29LVQNTYY551 pKa = 8.21QWVLKK556 pKa = 10.91NLVTLKK562 pKa = 10.93AQFLLGKK569 pKa = 9.96EE570 pKa = 5.88DD571 pKa = 3.54MFQWDD576 pKa = 3.33AFEE579 pKa = 4.66AFEE582 pKa = 5.18SIIPQKK588 pKa = 10.32MAGQYY593 pKa = 10.54SGFARR598 pKa = 11.84AVLKK602 pKa = 9.91QMRR605 pKa = 11.84DD606 pKa = 3.47QEE608 pKa = 4.51VMKK611 pKa = 10.1TDD613 pKa = 3.47QFIKK617 pKa = 10.67LLPFCFSPPKK627 pKa = 10.06LRR629 pKa = 11.84SNGEE633 pKa = 3.67PYY635 pKa = 10.49QFLRR639 pKa = 11.84LILKK643 pKa = 10.43GGGEE647 pKa = 4.11NFIEE651 pKa = 4.26VRR653 pKa = 11.84KK654 pKa = 10.13GSPLFSYY661 pKa = 10.68NPQTEE666 pKa = 4.12VLTICGRR673 pKa = 11.84MMSLKK678 pKa = 10.66GKK680 pKa = 10.2IEE682 pKa = 4.01DD683 pKa = 3.72EE684 pKa = 3.95EE685 pKa = 5.12RR686 pKa = 11.84NRR688 pKa = 11.84SMGNAVLAGFLVSGKK703 pKa = 10.1YY704 pKa = 10.53DD705 pKa = 3.9PDD707 pKa = 5.06LGDD710 pKa = 3.6FKK712 pKa = 10.87TIEE715 pKa = 4.29EE716 pKa = 4.4LEE718 pKa = 4.1KK719 pKa = 11.0LKK721 pKa = 10.75PGEE724 pKa = 3.97KK725 pKa = 10.57ANILLYY731 pKa = 10.07QGKK734 pKa = 7.97PVKK737 pKa = 9.66VVKK740 pKa = 10.37RR741 pKa = 11.84KK742 pKa = 9.92RR743 pKa = 11.84YY744 pKa = 8.87SALSNDD750 pKa = 2.67ISQGIKK756 pKa = 8.29RR757 pKa = 11.84QRR759 pKa = 11.84MTVEE763 pKa = 3.77SMGWALSS770 pKa = 3.61
Molecular weight: 87.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4034 |
100 |
770 |
448.2 |
50.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.049 ± 0.429 | 2.256 ± 0.339 |
4.611 ± 0.268 | 7.313 ± 0.742 |
3.718 ± 0.296 | 7.362 ± 0.439 |
1.71 ± 0.309 | 6.619 ± 0.419 |
7.536 ± 0.368 | 9.147 ± 0.421 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.743 ± 0.407 | 5.429 ± 0.593 |
4.09 ± 0.359 | 3.074 ± 0.396 |
5.181 ± 0.408 | 6.544 ± 0.392 |
6.197 ± 0.53 | 5.231 ± 0.329 |
1.264 ± 0.219 | 2.925 ± 0.311 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
