
Chitinophaga sp. YR573
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Chitinophagia; Chitinophagales; Chitinophagaceae; Chitinophaga; unclassified Chitinophaga
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
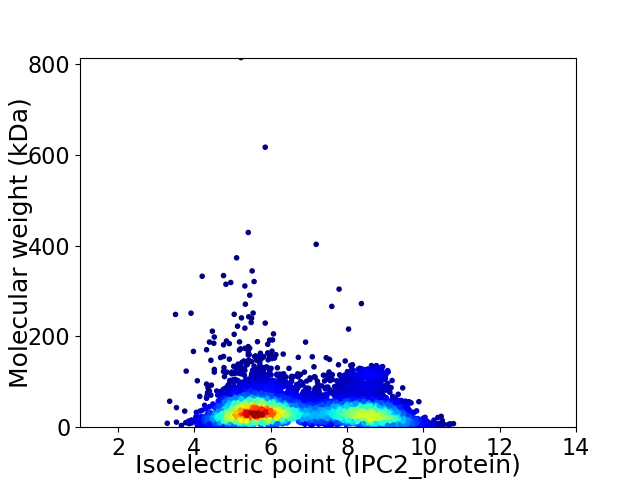
Virtual 2D-PAGE plot for 6483 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I0RVF3|A0A1I0RVF3_9BACT Uncharacterized protein OS=Chitinophaga sp. YR573 OX=1881040 GN=SAMN05428988_6014 PE=4 SV=1
MM1 pKa = 7.34YY2 pKa = 10.9NLTTSTALMKK12 pKa = 10.08NQKK15 pKa = 9.33AAQVLSDD22 pKa = 3.35QSLFSVLQNSQGQSLFFSIGDD43 pKa = 3.69DD44 pKa = 4.04AILYY48 pKa = 10.1LSAEE52 pKa = 4.2QDD54 pKa = 3.22NATTGWTPIDD64 pKa = 3.96LTTEE68 pKa = 4.2LAADD72 pKa = 3.83YY73 pKa = 9.62TGKK76 pKa = 8.3TVTAKK81 pKa = 10.07TFVVSQDD88 pKa = 2.88ATTGDD93 pKa = 3.7ILILQAIHH101 pKa = 5.84VAEE104 pKa = 4.99DD105 pKa = 3.39NADD108 pKa = 3.72YY109 pKa = 11.1LYY111 pKa = 10.42MLSGLSNAPDD121 pKa = 4.09AAWLTASSNRR131 pKa = 11.84SWTARR136 pKa = 11.84LYY138 pKa = 11.11DD139 pKa = 3.75DD140 pKa = 4.04TADD143 pKa = 3.75PVSNTDD149 pKa = 2.7IAYY152 pKa = 9.82INIALNQDD160 pKa = 4.84PIQAPYY166 pKa = 10.4VIVGLQDD173 pKa = 3.09QATTFIQNYY182 pKa = 7.77VVSIDD187 pKa = 3.46PTTTTGIWTKK197 pKa = 11.13YY198 pKa = 6.48EE199 pKa = 3.71TAEE202 pKa = 5.11NYY204 pKa = 8.99DD205 pKa = 3.48QLYY208 pKa = 10.76GMAIGKK214 pKa = 8.81AQAAMFAGLYY224 pKa = 9.47EE225 pKa = 5.39LYY227 pKa = 9.95TLNDD231 pKa = 3.45NTSLTFTPLKK241 pKa = 10.72SVFGPPDD248 pKa = 4.0IIKK251 pKa = 8.46LTPPDD256 pKa = 3.8GATAIATMPPDD267 pKa = 3.35SSGNTNLYY275 pKa = 9.66VAGTGAIYY283 pKa = 10.45LFSPDD288 pKa = 3.37QQLNDD293 pKa = 3.48ATGTAVITNDD303 pKa = 3.94LIVGVEE309 pKa = 4.04YY310 pKa = 10.5FKK312 pKa = 11.2VHH314 pKa = 5.51QSGNQVTLWGKK325 pKa = 10.27DD326 pKa = 3.2NEE328 pKa = 4.49GTVFYY333 pKa = 10.91SRR335 pKa = 11.84CDD337 pKa = 3.13VDD339 pKa = 4.16NQTDD343 pKa = 3.82PGSWSCPLPLLVQVEE358 pKa = 4.35QLASLLDD365 pKa = 3.5IQTQSSVLYY374 pKa = 10.58AHH376 pKa = 6.25TTGQSLVKK384 pKa = 10.36LIQDD388 pKa = 3.44AVTTQWSSQSILLPALSTSDD408 pKa = 2.71IYY410 pKa = 11.05EE411 pKa = 4.11YY412 pKa = 11.45YY413 pKa = 11.19AFTTTLKK420 pKa = 9.66VTDD423 pKa = 4.97DD424 pKa = 3.75NNLPIGGQAVQITATSPCSLYY445 pKa = 10.02MDD447 pKa = 3.84NQYY450 pKa = 8.53VTLLPDD456 pKa = 3.22VPLTVTSDD464 pKa = 3.36STGMITFVQEE474 pKa = 4.12TQTLGAVCYY483 pKa = 10.36NFQQDD488 pKa = 3.68DD489 pKa = 4.24NSWLNVNPMNNIVQSMSGITQGSDD513 pKa = 2.71LTAVNVTDD521 pKa = 4.35EE522 pKa = 4.53YY523 pKa = 11.26GTTTPLVPSTIPADD537 pKa = 3.65QADD540 pKa = 3.96AVAKK544 pKa = 10.62GIAQFVSTAQTLPQDD559 pKa = 3.88GSLQSTGSFAASEE572 pKa = 4.19GLVWGLSFANNSIQYY587 pKa = 10.14FEE589 pKa = 5.29GSSQYY594 pKa = 10.91AAAGIIPASDD604 pKa = 2.8IGNAIEE610 pKa = 5.38AFAGDD615 pKa = 3.26IFKK618 pKa = 10.22WLEE621 pKa = 3.74NAVDD625 pKa = 3.97EE626 pKa = 4.58VAQFVVKK633 pKa = 10.89VIDD636 pKa = 4.08GVTHH640 pKa = 6.08FFIEE644 pKa = 4.03IGDD647 pKa = 3.97AVYY650 pKa = 10.55HH651 pKa = 7.08FIMQCINDD659 pKa = 4.0VVHH662 pKa = 7.28GIQFILSKK670 pKa = 10.57IEE672 pKa = 3.88AGFEE676 pKa = 4.39KK677 pKa = 10.46LVQWVGFIFSWSDD690 pKa = 3.15IIRR693 pKa = 11.84THH695 pKa = 6.22NVLKK699 pKa = 10.95NIFAQYY705 pKa = 10.42SQYY708 pKa = 11.06CVDD711 pKa = 4.6NISSYY716 pKa = 10.96KK717 pKa = 8.69QTLLDD722 pKa = 3.57TLSTINEE729 pKa = 4.93EE730 pKa = 3.5IDD732 pKa = 2.93QWANIPAVSDD742 pKa = 4.08TIAGSNASSPSMPGKK757 pKa = 10.22DD758 pKa = 3.13SPQSHH763 pKa = 6.26WALYY767 pKa = 7.29HH768 pKa = 5.43TKK770 pKa = 10.61GNVAGASTTFDD781 pKa = 3.0ASFDD785 pKa = 3.73GDD787 pKa = 3.96ALSLIIADD795 pKa = 4.3LCTALEE801 pKa = 4.06NEE803 pKa = 4.36EE804 pKa = 4.34QNFEE808 pKa = 4.29DD809 pKa = 4.36VLAALKK815 pKa = 9.98TDD817 pKa = 3.92VIDD820 pKa = 4.69QIGTLSASEE829 pKa = 5.27LITNLLGIIANLLIDD844 pKa = 3.76TVGNIIGTVMDD855 pKa = 4.99IIAALVAGVVEE866 pKa = 4.88ALTTPLDD873 pKa = 3.61IPVISYY879 pKa = 9.95IYY881 pKa = 11.06NKK883 pKa = 9.13FTGNDD888 pKa = 3.4LTLLDD893 pKa = 4.42LTCLIVSIPVTIIYY907 pKa = 10.34KK908 pKa = 10.04LIKK911 pKa = 9.86EE912 pKa = 4.34ATPFPDD918 pKa = 5.1DD919 pKa = 4.67SFTQSLIAATDD930 pKa = 3.38YY931 pKa = 7.9TTIQNLYY938 pKa = 10.21AGASSLAVADD948 pKa = 4.64DD949 pKa = 4.69DD950 pKa = 5.11GVTPVQIITLTLAISGYY967 pKa = 9.46FGSIAMIIMTAVKK980 pKa = 10.04AAAPQSAAVCTVYY993 pKa = 11.24ALAYY997 pKa = 9.97LPYY1000 pKa = 9.39IAPNIAITKK1009 pKa = 9.62PSTQKK1014 pKa = 10.07WYY1016 pKa = 11.1VDD1018 pKa = 3.25MDD1020 pKa = 4.23EE1021 pKa = 5.13IITVISVIKK1030 pKa = 9.26TFSDD1034 pKa = 2.82ITLYY1038 pKa = 10.59KK1039 pKa = 9.84YY1040 pKa = 10.91DD1041 pKa = 4.28KK1042 pKa = 10.34NDD1044 pKa = 3.67PEE1046 pKa = 5.13ASQTLAKK1053 pKa = 8.61WAKK1056 pKa = 8.62ASPYY1060 pKa = 10.32VEE1062 pKa = 4.91LLINVIWEE1070 pKa = 3.85IPTIGFIADD1079 pKa = 3.5NQDD1082 pKa = 2.74AGTVLGFLGNTSFNVSGILSPWSDD1106 pKa = 2.98TDD1108 pKa = 3.7YY1109 pKa = 11.88VMLGIVIFIGLYY1121 pKa = 9.81GEE1123 pKa = 4.2MMLVQGMVNFPAPSSAKK1140 pKa = 10.15YY1141 pKa = 10.12LATKK1145 pKa = 9.51EE1146 pKa = 4.02AA1147 pKa = 3.95
MM1 pKa = 7.34YY2 pKa = 10.9NLTTSTALMKK12 pKa = 10.08NQKK15 pKa = 9.33AAQVLSDD22 pKa = 3.35QSLFSVLQNSQGQSLFFSIGDD43 pKa = 3.69DD44 pKa = 4.04AILYY48 pKa = 10.1LSAEE52 pKa = 4.2QDD54 pKa = 3.22NATTGWTPIDD64 pKa = 3.96LTTEE68 pKa = 4.2LAADD72 pKa = 3.83YY73 pKa = 9.62TGKK76 pKa = 8.3TVTAKK81 pKa = 10.07TFVVSQDD88 pKa = 2.88ATTGDD93 pKa = 3.7ILILQAIHH101 pKa = 5.84VAEE104 pKa = 4.99DD105 pKa = 3.39NADD108 pKa = 3.72YY109 pKa = 11.1LYY111 pKa = 10.42MLSGLSNAPDD121 pKa = 4.09AAWLTASSNRR131 pKa = 11.84SWTARR136 pKa = 11.84LYY138 pKa = 11.11DD139 pKa = 3.75DD140 pKa = 4.04TADD143 pKa = 3.75PVSNTDD149 pKa = 2.7IAYY152 pKa = 9.82INIALNQDD160 pKa = 4.84PIQAPYY166 pKa = 10.4VIVGLQDD173 pKa = 3.09QATTFIQNYY182 pKa = 7.77VVSIDD187 pKa = 3.46PTTTTGIWTKK197 pKa = 11.13YY198 pKa = 6.48EE199 pKa = 3.71TAEE202 pKa = 5.11NYY204 pKa = 8.99DD205 pKa = 3.48QLYY208 pKa = 10.76GMAIGKK214 pKa = 8.81AQAAMFAGLYY224 pKa = 9.47EE225 pKa = 5.39LYY227 pKa = 9.95TLNDD231 pKa = 3.45NTSLTFTPLKK241 pKa = 10.72SVFGPPDD248 pKa = 4.0IIKK251 pKa = 8.46LTPPDD256 pKa = 3.8GATAIATMPPDD267 pKa = 3.35SSGNTNLYY275 pKa = 9.66VAGTGAIYY283 pKa = 10.45LFSPDD288 pKa = 3.37QQLNDD293 pKa = 3.48ATGTAVITNDD303 pKa = 3.94LIVGVEE309 pKa = 4.04YY310 pKa = 10.5FKK312 pKa = 11.2VHH314 pKa = 5.51QSGNQVTLWGKK325 pKa = 10.27DD326 pKa = 3.2NEE328 pKa = 4.49GTVFYY333 pKa = 10.91SRR335 pKa = 11.84CDD337 pKa = 3.13VDD339 pKa = 4.16NQTDD343 pKa = 3.82PGSWSCPLPLLVQVEE358 pKa = 4.35QLASLLDD365 pKa = 3.5IQTQSSVLYY374 pKa = 10.58AHH376 pKa = 6.25TTGQSLVKK384 pKa = 10.36LIQDD388 pKa = 3.44AVTTQWSSQSILLPALSTSDD408 pKa = 2.71IYY410 pKa = 11.05EE411 pKa = 4.11YY412 pKa = 11.45YY413 pKa = 11.19AFTTTLKK420 pKa = 9.66VTDD423 pKa = 4.97DD424 pKa = 3.75NNLPIGGQAVQITATSPCSLYY445 pKa = 10.02MDD447 pKa = 3.84NQYY450 pKa = 8.53VTLLPDD456 pKa = 3.22VPLTVTSDD464 pKa = 3.36STGMITFVQEE474 pKa = 4.12TQTLGAVCYY483 pKa = 10.36NFQQDD488 pKa = 3.68DD489 pKa = 4.24NSWLNVNPMNNIVQSMSGITQGSDD513 pKa = 2.71LTAVNVTDD521 pKa = 4.35EE522 pKa = 4.53YY523 pKa = 11.26GTTTPLVPSTIPADD537 pKa = 3.65QADD540 pKa = 3.96AVAKK544 pKa = 10.62GIAQFVSTAQTLPQDD559 pKa = 3.88GSLQSTGSFAASEE572 pKa = 4.19GLVWGLSFANNSIQYY587 pKa = 10.14FEE589 pKa = 5.29GSSQYY594 pKa = 10.91AAAGIIPASDD604 pKa = 2.8IGNAIEE610 pKa = 5.38AFAGDD615 pKa = 3.26IFKK618 pKa = 10.22WLEE621 pKa = 3.74NAVDD625 pKa = 3.97EE626 pKa = 4.58VAQFVVKK633 pKa = 10.89VIDD636 pKa = 4.08GVTHH640 pKa = 6.08FFIEE644 pKa = 4.03IGDD647 pKa = 3.97AVYY650 pKa = 10.55HH651 pKa = 7.08FIMQCINDD659 pKa = 4.0VVHH662 pKa = 7.28GIQFILSKK670 pKa = 10.57IEE672 pKa = 3.88AGFEE676 pKa = 4.39KK677 pKa = 10.46LVQWVGFIFSWSDD690 pKa = 3.15IIRR693 pKa = 11.84THH695 pKa = 6.22NVLKK699 pKa = 10.95NIFAQYY705 pKa = 10.42SQYY708 pKa = 11.06CVDD711 pKa = 4.6NISSYY716 pKa = 10.96KK717 pKa = 8.69QTLLDD722 pKa = 3.57TLSTINEE729 pKa = 4.93EE730 pKa = 3.5IDD732 pKa = 2.93QWANIPAVSDD742 pKa = 4.08TIAGSNASSPSMPGKK757 pKa = 10.22DD758 pKa = 3.13SPQSHH763 pKa = 6.26WALYY767 pKa = 7.29HH768 pKa = 5.43TKK770 pKa = 10.61GNVAGASTTFDD781 pKa = 3.0ASFDD785 pKa = 3.73GDD787 pKa = 3.96ALSLIIADD795 pKa = 4.3LCTALEE801 pKa = 4.06NEE803 pKa = 4.36EE804 pKa = 4.34QNFEE808 pKa = 4.29DD809 pKa = 4.36VLAALKK815 pKa = 9.98TDD817 pKa = 3.92VIDD820 pKa = 4.69QIGTLSASEE829 pKa = 5.27LITNLLGIIANLLIDD844 pKa = 3.76TVGNIIGTVMDD855 pKa = 4.99IIAALVAGVVEE866 pKa = 4.88ALTTPLDD873 pKa = 3.61IPVISYY879 pKa = 9.95IYY881 pKa = 11.06NKK883 pKa = 9.13FTGNDD888 pKa = 3.4LTLLDD893 pKa = 4.42LTCLIVSIPVTIIYY907 pKa = 10.34KK908 pKa = 10.04LIKK911 pKa = 9.86EE912 pKa = 4.34ATPFPDD918 pKa = 5.1DD919 pKa = 4.67SFTQSLIAATDD930 pKa = 3.38YY931 pKa = 7.9TTIQNLYY938 pKa = 10.21AGASSLAVADD948 pKa = 4.64DD949 pKa = 4.69DD950 pKa = 5.11GVTPVQIITLTLAISGYY967 pKa = 9.46FGSIAMIIMTAVKK980 pKa = 10.04AAAPQSAAVCTVYY993 pKa = 11.24ALAYY997 pKa = 9.97LPYY1000 pKa = 9.39IAPNIAITKK1009 pKa = 9.62PSTQKK1014 pKa = 10.07WYY1016 pKa = 11.1VDD1018 pKa = 3.25MDD1020 pKa = 4.23EE1021 pKa = 5.13IITVISVIKK1030 pKa = 9.26TFSDD1034 pKa = 2.82ITLYY1038 pKa = 10.59KK1039 pKa = 9.84YY1040 pKa = 10.91DD1041 pKa = 4.28KK1042 pKa = 10.34NDD1044 pKa = 3.67PEE1046 pKa = 5.13ASQTLAKK1053 pKa = 8.61WAKK1056 pKa = 8.62ASPYY1060 pKa = 10.32VEE1062 pKa = 4.91LLINVIWEE1070 pKa = 3.85IPTIGFIADD1079 pKa = 3.5NQDD1082 pKa = 2.74AGTVLGFLGNTSFNVSGILSPWSDD1106 pKa = 2.98TDD1108 pKa = 3.7YY1109 pKa = 11.88VMLGIVIFIGLYY1121 pKa = 9.81GEE1123 pKa = 4.2MMLVQGMVNFPAPSSAKK1140 pKa = 10.15YY1141 pKa = 10.12LATKK1145 pKa = 9.51EE1146 pKa = 4.02AA1147 pKa = 3.95
Molecular weight: 123.64 kDa
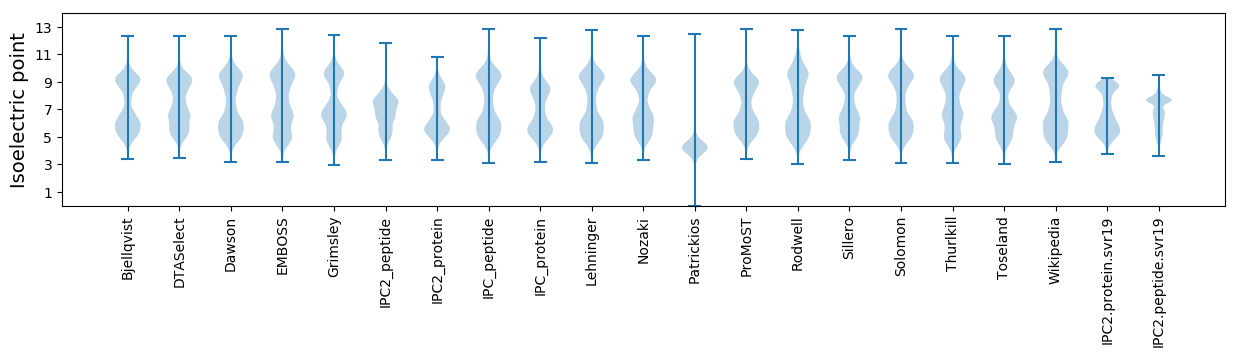
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I0Q2Q1|A0A1I0Q2Q1_9BACT Uncharacterized protein OS=Chitinophaga sp. YR573 OX=1881040 GN=SAMN05428988_3166 PE=4 SV=1
MM1 pKa = 6.41NTKK4 pKa = 9.41IRR6 pKa = 11.84AFIIMLLLYY15 pKa = 9.7GLHH18 pKa = 6.08VYY20 pKa = 10.76LMNTLLSIHH29 pKa = 6.78SNMSDD34 pKa = 2.46ILAFLMMIACPFYY47 pKa = 10.97KK48 pKa = 9.84RR49 pKa = 11.84IHH51 pKa = 5.53QRR53 pKa = 11.84LVATLKK59 pKa = 10.66NRR61 pKa = 3.55
MM1 pKa = 6.41NTKK4 pKa = 9.41IRR6 pKa = 11.84AFIIMLLLYY15 pKa = 9.7GLHH18 pKa = 6.08VYY20 pKa = 10.76LMNTLLSIHH29 pKa = 6.78SNMSDD34 pKa = 2.46ILAFLMMIACPFYY47 pKa = 10.97KK48 pKa = 9.84RR49 pKa = 11.84IHH51 pKa = 5.53QRR53 pKa = 11.84LVATLKK59 pKa = 10.66NRR61 pKa = 3.55
Molecular weight: 7.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2436871 |
29 |
7223 |
375.9 |
42.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.613 ± 0.033 | 0.826 ± 0.011 |
5.334 ± 0.021 | 5.302 ± 0.036 |
4.577 ± 0.021 | 6.936 ± 0.029 |
1.92 ± 0.015 | 7.016 ± 0.027 |
6.352 ± 0.029 | 9.389 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.337 ± 0.016 | 5.499 ± 0.029 |
4.023 ± 0.021 | 3.89 ± 0.021 |
4.007 ± 0.022 | 6.466 ± 0.031 |
6.428 ± 0.058 | 6.436 ± 0.028 |
1.297 ± 0.013 | 4.352 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
