
Hydrocarboniphaga daqingensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Nevskiales; Sinobacteraceae; Hydrocarboniphaga
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
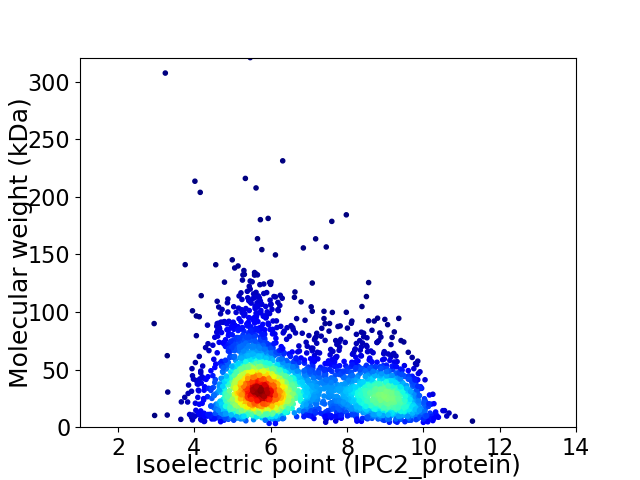
Virtual 2D-PAGE plot for 3253 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M5LZQ8|A0A1M5LZQ8_9GAMM NAD(P)-dependent dehydrogenase short-chain alcohol dehydrogenase family OS=Hydrocarboniphaga daqingensis OX=490188 GN=SAMN04488068_1170 PE=3 SV=1
MM1 pKa = 7.4KK2 pKa = 9.68MHH4 pKa = 6.6YY5 pKa = 9.35LAAAAAVFGCTTASAATLADD25 pKa = 3.63LAKK28 pKa = 10.36EE29 pKa = 4.25VTASGWISGSYY40 pKa = 10.2AYY42 pKa = 10.79NFNDD46 pKa = 4.1SNGLIQGRR54 pKa = 11.84VADD57 pKa = 3.91VQADD61 pKa = 3.99SFVLNQAVLNLSSAPTEE78 pKa = 4.36GFGGTVMLLAGEE90 pKa = 4.46DD91 pKa = 3.46AANVINPSYY100 pKa = 11.33GDD102 pKa = 3.57TDD104 pKa = 3.81DD105 pKa = 5.11KK106 pKa = 11.58LAVQQAYY113 pKa = 10.45LSYY116 pKa = 10.85AAGDD120 pKa = 3.64LTVIAGRR127 pKa = 11.84YY128 pKa = 8.48NSLAGYY134 pKa = 8.47EE135 pKa = 4.19VLADD139 pKa = 4.06PSNTTLSRR147 pKa = 11.84SLVYY151 pKa = 10.45INAQPYY157 pKa = 8.03FHH159 pKa = 6.72TGVRR163 pKa = 11.84AAYY166 pKa = 8.63KK167 pKa = 10.6ASDD170 pKa = 3.43AATLFLGLANSAALGASVDD189 pKa = 3.79ANDD192 pKa = 4.18PKK194 pKa = 10.53TIEE197 pKa = 3.99VGGLFTLSPAVTVGIYY213 pKa = 10.21DD214 pKa = 3.71YY215 pKa = 11.17FGVEE219 pKa = 4.8AGDD222 pKa = 3.43TGKK225 pKa = 8.52NTNYY229 pKa = 10.57LDD231 pKa = 3.89FVGTLTATDD240 pKa = 3.74TLSFAINADD249 pKa = 3.64YY250 pKa = 10.44FSQEE254 pKa = 3.83DD255 pKa = 4.05TVDD258 pKa = 3.56VVGVAGYY265 pKa = 7.61ATLKK269 pKa = 10.82LNDD272 pKa = 3.38MWSATVRR279 pKa = 11.84LEE281 pKa = 4.1NVNIEE286 pKa = 4.03PDD288 pKa = 3.39AGSDD292 pKa = 3.46LTINEE297 pKa = 3.88ATLALTYY304 pKa = 10.88APASNFKK311 pKa = 9.78MFFEE315 pKa = 4.7GRR317 pKa = 11.84VDD319 pKa = 4.85DD320 pKa = 5.3ADD322 pKa = 4.01ADD324 pKa = 3.84IYY326 pKa = 10.93IDD328 pKa = 3.69GTDD331 pKa = 3.81FTGTQPTLGVKK342 pKa = 10.36AIYY345 pKa = 9.12TFGMM349 pKa = 3.94
MM1 pKa = 7.4KK2 pKa = 9.68MHH4 pKa = 6.6YY5 pKa = 9.35LAAAAAVFGCTTASAATLADD25 pKa = 3.63LAKK28 pKa = 10.36EE29 pKa = 4.25VTASGWISGSYY40 pKa = 10.2AYY42 pKa = 10.79NFNDD46 pKa = 4.1SNGLIQGRR54 pKa = 11.84VADD57 pKa = 3.91VQADD61 pKa = 3.99SFVLNQAVLNLSSAPTEE78 pKa = 4.36GFGGTVMLLAGEE90 pKa = 4.46DD91 pKa = 3.46AANVINPSYY100 pKa = 11.33GDD102 pKa = 3.57TDD104 pKa = 3.81DD105 pKa = 5.11KK106 pKa = 11.58LAVQQAYY113 pKa = 10.45LSYY116 pKa = 10.85AAGDD120 pKa = 3.64LTVIAGRR127 pKa = 11.84YY128 pKa = 8.48NSLAGYY134 pKa = 8.47EE135 pKa = 4.19VLADD139 pKa = 4.06PSNTTLSRR147 pKa = 11.84SLVYY151 pKa = 10.45INAQPYY157 pKa = 8.03FHH159 pKa = 6.72TGVRR163 pKa = 11.84AAYY166 pKa = 8.63KK167 pKa = 10.6ASDD170 pKa = 3.43AATLFLGLANSAALGASVDD189 pKa = 3.79ANDD192 pKa = 4.18PKK194 pKa = 10.53TIEE197 pKa = 3.99VGGLFTLSPAVTVGIYY213 pKa = 10.21DD214 pKa = 3.71YY215 pKa = 11.17FGVEE219 pKa = 4.8AGDD222 pKa = 3.43TGKK225 pKa = 8.52NTNYY229 pKa = 10.57LDD231 pKa = 3.89FVGTLTATDD240 pKa = 3.74TLSFAINADD249 pKa = 3.64YY250 pKa = 10.44FSQEE254 pKa = 3.83DD255 pKa = 4.05TVDD258 pKa = 3.56VVGVAGYY265 pKa = 7.61ATLKK269 pKa = 10.82LNDD272 pKa = 3.38MWSATVRR279 pKa = 11.84LEE281 pKa = 4.1NVNIEE286 pKa = 4.03PDD288 pKa = 3.39AGSDD292 pKa = 3.46LTINEE297 pKa = 3.88ATLALTYY304 pKa = 10.88APASNFKK311 pKa = 9.78MFFEE315 pKa = 4.7GRR317 pKa = 11.84VDD319 pKa = 4.85DD320 pKa = 5.3ADD322 pKa = 4.01ADD324 pKa = 3.84IYY326 pKa = 10.93IDD328 pKa = 3.69GTDD331 pKa = 3.81FTGTQPTLGVKK342 pKa = 10.36AIYY345 pKa = 9.12TFGMM349 pKa = 3.94
Molecular weight: 36.58 kDa
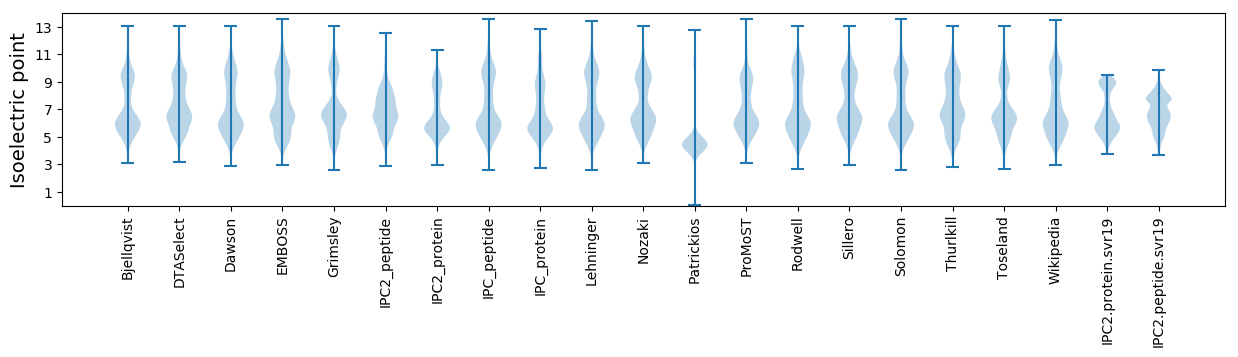
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M5LJC7|A0A1M5LJC7_9GAMM Exopolyphosphatase / guanosine-5'-triphosphate 3'-diphosphate pyrophosphatase OS=Hydrocarboniphaga daqingensis OX=490188 GN=SAMN04488068_0930 PE=4 SV=1
MM1 pKa = 7.38KK2 pKa = 9.54RR3 pKa = 11.84TFQPHH8 pKa = 4.47SLRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 7.45RR14 pKa = 11.84THH16 pKa = 6.03GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.01VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.32GRR39 pKa = 11.84ARR41 pKa = 11.84LTPVV45 pKa = 2.71
MM1 pKa = 7.38KK2 pKa = 9.54RR3 pKa = 11.84TFQPHH8 pKa = 4.47SLRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 7.45RR14 pKa = 11.84THH16 pKa = 6.03GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.01VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.32GRR39 pKa = 11.84ARR41 pKa = 11.84LTPVV45 pKa = 2.71
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1160950 |
28 |
3144 |
356.9 |
38.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.608 ± 0.059 | 0.886 ± 0.012 |
6.016 ± 0.035 | 4.862 ± 0.037 |
3.29 ± 0.026 | 8.208 ± 0.046 |
2.075 ± 0.022 | 4.566 ± 0.028 |
2.717 ± 0.038 | 10.904 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.169 ± 0.021 | 2.607 ± 0.027 |
5.379 ± 0.033 | 4.04 ± 0.026 |
7.552 ± 0.045 | 5.661 ± 0.036 |
5.176 ± 0.038 | 7.375 ± 0.034 |
1.386 ± 0.018 | 2.523 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
