
Kigluaik phantom orthophasmavirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phasmaviridae; Orthophasmavirus
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
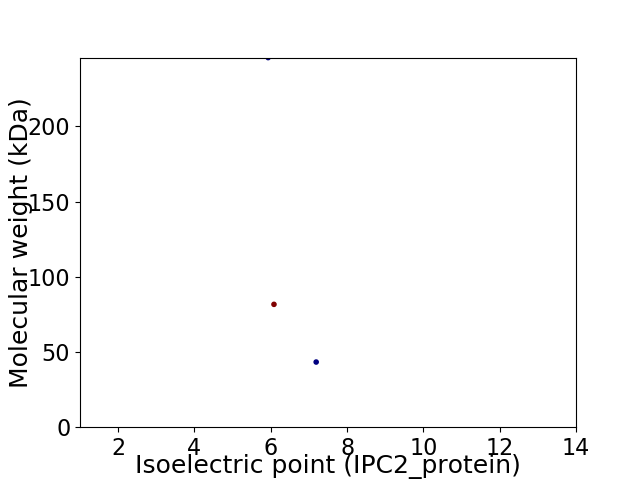
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A059XT43|A0A059XT43_9VIRU Nucleoprotein OS=Kigluaik phantom orthophasmavirus OX=1980539 GN=N PE=4 SV=1
MM1 pKa = 6.8KK2 pKa = 10.46QKK4 pKa = 10.4IGTNMLMFLFFYY16 pKa = 10.66NIIIAEE22 pKa = 4.44GSHH25 pKa = 6.3GPNIYY30 pKa = 10.29SKK32 pKa = 10.34NGEE35 pKa = 4.21VEE37 pKa = 4.48FIGYY41 pKa = 8.4DD42 pKa = 3.2EE43 pKa = 5.49CSISVNKK50 pKa = 9.54MVINSTNGIFPGIHH64 pKa = 6.51FGLIEE69 pKa = 4.1YY70 pKa = 8.82TCDD73 pKa = 3.65KK74 pKa = 9.74ITGKK78 pKa = 10.22IISRR82 pKa = 11.84MSCMEE87 pKa = 3.47CRR89 pKa = 11.84FYY91 pKa = 10.69CTHH94 pKa = 5.98NQNIVEE100 pKa = 4.25CSNYY104 pKa = 10.09YY105 pKa = 9.91IPLISGFSVGAIFMIILIYY124 pKa = 10.49VLSKK128 pKa = 11.13NDD130 pKa = 3.21VLKK133 pKa = 10.84RR134 pKa = 11.84ISNSLEE140 pKa = 3.63NNVISKK146 pKa = 10.58LSSRR150 pKa = 11.84KK151 pKa = 9.38SKK153 pKa = 10.23NARR156 pKa = 11.84RR157 pKa = 11.84LLKK160 pKa = 10.29KK161 pKa = 10.23IRR163 pKa = 11.84DD164 pKa = 3.51AEE166 pKa = 4.53AINNSNIKK174 pKa = 9.79MIGSTGQEE182 pKa = 4.43DD183 pKa = 3.57IDD185 pKa = 4.88LHH187 pKa = 8.35NEE189 pKa = 3.95DD190 pKa = 4.75DD191 pKa = 4.07NKK193 pKa = 11.45GIMKK197 pKa = 10.42RR198 pKa = 11.84GLNVEE203 pKa = 4.33ASNDD207 pKa = 2.9IDD209 pKa = 4.22IGSNSNGHH217 pKa = 6.24LFYY220 pKa = 11.23YY221 pKa = 9.43EE222 pKa = 4.66HH223 pKa = 7.09EE224 pKa = 4.79PEE226 pKa = 5.82AIEE229 pKa = 3.96YY230 pKa = 9.47KK231 pKa = 10.4RR232 pKa = 11.84RR233 pKa = 11.84GSVPRR238 pKa = 11.84NVTILSMIIMLSIPPLILGCDD259 pKa = 2.87NTFFLNSEE267 pKa = 4.39GKK269 pKa = 8.65VCDD272 pKa = 4.26YY273 pKa = 10.87KK274 pKa = 11.39GCQNKK279 pKa = 9.24NFVSMPLMKK288 pKa = 10.46GNRR291 pKa = 11.84ICVRR295 pKa = 11.84DD296 pKa = 3.48SDD298 pKa = 4.19QNILAISITEE308 pKa = 3.82SNIIYY313 pKa = 9.71RR314 pKa = 11.84HH315 pKa = 5.51HH316 pKa = 7.29LMYY319 pKa = 10.16YY320 pKa = 6.19TTDD323 pKa = 3.33YY324 pKa = 11.09EE325 pKa = 4.48IDD327 pKa = 3.85VKK329 pKa = 11.2HH330 pKa = 5.92HH331 pKa = 5.66TNCLGAGKK339 pKa = 10.12CDD341 pKa = 3.54EE342 pKa = 4.67EE343 pKa = 4.39KK344 pKa = 11.1CNLISDD350 pKa = 4.64EE351 pKa = 4.41YY352 pKa = 11.47KK353 pKa = 11.27GMIPNNSVSGNGCSEE368 pKa = 3.86GFDD371 pKa = 3.43RR372 pKa = 11.84CSNMCFYY379 pKa = 11.0SVSCTFYY386 pKa = 10.61SWYY389 pKa = 10.56LKK391 pKa = 10.12EE392 pKa = 5.04GKK394 pKa = 9.2IKK396 pKa = 10.71YY397 pKa = 8.73PVYY400 pKa = 10.66TMSSQNWEE408 pKa = 3.54VSIAFEE414 pKa = 4.15YY415 pKa = 10.84LGARR419 pKa = 11.84YY420 pKa = 9.45IKK422 pKa = 10.38ILDD425 pKa = 3.55VNNPDD430 pKa = 3.22GSIMINGKK438 pKa = 7.28EE439 pKa = 3.95VKK441 pKa = 10.03IGISSFTSSSIRR453 pKa = 11.84LEE455 pKa = 3.88RR456 pKa = 11.84DD457 pKa = 3.16SIIINNTLYY466 pKa = 11.12LVDD469 pKa = 4.15SSFANMPEE477 pKa = 3.75TDD479 pKa = 4.58KK480 pKa = 11.5IGDD483 pKa = 3.64YY484 pKa = 10.83QLAIDD489 pKa = 4.54GKK491 pKa = 8.53SRR493 pKa = 11.84VYY495 pKa = 8.61NTHH498 pKa = 7.16NIRR501 pKa = 11.84CQTDD505 pKa = 3.78FCKK508 pKa = 8.48TTCASPEE515 pKa = 3.78PKK517 pKa = 9.59INRR520 pKa = 11.84FISSIEE526 pKa = 3.9RR527 pKa = 11.84YY528 pKa = 9.54RR529 pKa = 11.84SLKK532 pKa = 9.73FSKK535 pKa = 10.07IGNSNNINVHH545 pKa = 5.43EE546 pKa = 4.48PVNGLITMNIGDD558 pKa = 4.06IRR560 pKa = 11.84IEE562 pKa = 3.96SLQVIDD568 pKa = 4.69ASCDD572 pKa = 3.07IEE574 pKa = 5.84VMMTYY579 pKa = 10.31SCVGCDD585 pKa = 3.07VDD587 pKa = 5.37SYY589 pKa = 12.08AVVQAYY595 pKa = 9.71NIKK598 pKa = 10.09QSGIIPITSTCNLVKK613 pKa = 10.44DD614 pKa = 4.38YY615 pKa = 10.71ISCEE619 pKa = 3.69DD620 pKa = 4.22TINKK624 pKa = 9.35IEE626 pKa = 4.19FVDD629 pKa = 3.78NSKK632 pKa = 10.39VCYY635 pKa = 10.11ISMKK639 pKa = 10.03GRR641 pKa = 11.84NDD643 pKa = 3.62TIKK646 pKa = 10.7IMFNTTYY653 pKa = 9.91LGSMDD658 pKa = 4.08PSKK661 pKa = 10.94SIYY664 pKa = 9.03ATSSRR669 pKa = 11.84MDD671 pKa = 5.38DD672 pKa = 2.83ITGLVTSSGFINGIISTIGSISLFTIIGTFATRR705 pKa = 11.84LIGVILTMRR714 pKa = 11.84SVDD717 pKa = 3.46KK718 pKa = 10.87EE719 pKa = 4.42SYY721 pKa = 9.92RR722 pKa = 11.84SNAIII727 pKa = 4.38
MM1 pKa = 6.8KK2 pKa = 10.46QKK4 pKa = 10.4IGTNMLMFLFFYY16 pKa = 10.66NIIIAEE22 pKa = 4.44GSHH25 pKa = 6.3GPNIYY30 pKa = 10.29SKK32 pKa = 10.34NGEE35 pKa = 4.21VEE37 pKa = 4.48FIGYY41 pKa = 8.4DD42 pKa = 3.2EE43 pKa = 5.49CSISVNKK50 pKa = 9.54MVINSTNGIFPGIHH64 pKa = 6.51FGLIEE69 pKa = 4.1YY70 pKa = 8.82TCDD73 pKa = 3.65KK74 pKa = 9.74ITGKK78 pKa = 10.22IISRR82 pKa = 11.84MSCMEE87 pKa = 3.47CRR89 pKa = 11.84FYY91 pKa = 10.69CTHH94 pKa = 5.98NQNIVEE100 pKa = 4.25CSNYY104 pKa = 10.09YY105 pKa = 9.91IPLISGFSVGAIFMIILIYY124 pKa = 10.49VLSKK128 pKa = 11.13NDD130 pKa = 3.21VLKK133 pKa = 10.84RR134 pKa = 11.84ISNSLEE140 pKa = 3.63NNVISKK146 pKa = 10.58LSSRR150 pKa = 11.84KK151 pKa = 9.38SKK153 pKa = 10.23NARR156 pKa = 11.84RR157 pKa = 11.84LLKK160 pKa = 10.29KK161 pKa = 10.23IRR163 pKa = 11.84DD164 pKa = 3.51AEE166 pKa = 4.53AINNSNIKK174 pKa = 9.79MIGSTGQEE182 pKa = 4.43DD183 pKa = 3.57IDD185 pKa = 4.88LHH187 pKa = 8.35NEE189 pKa = 3.95DD190 pKa = 4.75DD191 pKa = 4.07NKK193 pKa = 11.45GIMKK197 pKa = 10.42RR198 pKa = 11.84GLNVEE203 pKa = 4.33ASNDD207 pKa = 2.9IDD209 pKa = 4.22IGSNSNGHH217 pKa = 6.24LFYY220 pKa = 11.23YY221 pKa = 9.43EE222 pKa = 4.66HH223 pKa = 7.09EE224 pKa = 4.79PEE226 pKa = 5.82AIEE229 pKa = 3.96YY230 pKa = 9.47KK231 pKa = 10.4RR232 pKa = 11.84RR233 pKa = 11.84GSVPRR238 pKa = 11.84NVTILSMIIMLSIPPLILGCDD259 pKa = 2.87NTFFLNSEE267 pKa = 4.39GKK269 pKa = 8.65VCDD272 pKa = 4.26YY273 pKa = 10.87KK274 pKa = 11.39GCQNKK279 pKa = 9.24NFVSMPLMKK288 pKa = 10.46GNRR291 pKa = 11.84ICVRR295 pKa = 11.84DD296 pKa = 3.48SDD298 pKa = 4.19QNILAISITEE308 pKa = 3.82SNIIYY313 pKa = 9.71RR314 pKa = 11.84HH315 pKa = 5.51HH316 pKa = 7.29LMYY319 pKa = 10.16YY320 pKa = 6.19TTDD323 pKa = 3.33YY324 pKa = 11.09EE325 pKa = 4.48IDD327 pKa = 3.85VKK329 pKa = 11.2HH330 pKa = 5.92HH331 pKa = 5.66TNCLGAGKK339 pKa = 10.12CDD341 pKa = 3.54EE342 pKa = 4.67EE343 pKa = 4.39KK344 pKa = 11.1CNLISDD350 pKa = 4.64EE351 pKa = 4.41YY352 pKa = 11.47KK353 pKa = 11.27GMIPNNSVSGNGCSEE368 pKa = 3.86GFDD371 pKa = 3.43RR372 pKa = 11.84CSNMCFYY379 pKa = 11.0SVSCTFYY386 pKa = 10.61SWYY389 pKa = 10.56LKK391 pKa = 10.12EE392 pKa = 5.04GKK394 pKa = 9.2IKK396 pKa = 10.71YY397 pKa = 8.73PVYY400 pKa = 10.66TMSSQNWEE408 pKa = 3.54VSIAFEE414 pKa = 4.15YY415 pKa = 10.84LGARR419 pKa = 11.84YY420 pKa = 9.45IKK422 pKa = 10.38ILDD425 pKa = 3.55VNNPDD430 pKa = 3.22GSIMINGKK438 pKa = 7.28EE439 pKa = 3.95VKK441 pKa = 10.03IGISSFTSSSIRR453 pKa = 11.84LEE455 pKa = 3.88RR456 pKa = 11.84DD457 pKa = 3.16SIIINNTLYY466 pKa = 11.12LVDD469 pKa = 4.15SSFANMPEE477 pKa = 3.75TDD479 pKa = 4.58KK480 pKa = 11.5IGDD483 pKa = 3.64YY484 pKa = 10.83QLAIDD489 pKa = 4.54GKK491 pKa = 8.53SRR493 pKa = 11.84VYY495 pKa = 8.61NTHH498 pKa = 7.16NIRR501 pKa = 11.84CQTDD505 pKa = 3.78FCKK508 pKa = 8.48TTCASPEE515 pKa = 3.78PKK517 pKa = 9.59INRR520 pKa = 11.84FISSIEE526 pKa = 3.9RR527 pKa = 11.84YY528 pKa = 9.54RR529 pKa = 11.84SLKK532 pKa = 9.73FSKK535 pKa = 10.07IGNSNNINVHH545 pKa = 5.43EE546 pKa = 4.48PVNGLITMNIGDD558 pKa = 4.06IRR560 pKa = 11.84IEE562 pKa = 3.96SLQVIDD568 pKa = 4.69ASCDD572 pKa = 3.07IEE574 pKa = 5.84VMMTYY579 pKa = 10.31SCVGCDD585 pKa = 3.07VDD587 pKa = 5.37SYY589 pKa = 12.08AVVQAYY595 pKa = 9.71NIKK598 pKa = 10.09QSGIIPITSTCNLVKK613 pKa = 10.44DD614 pKa = 4.38YY615 pKa = 10.71ISCEE619 pKa = 3.69DD620 pKa = 4.22TINKK624 pKa = 9.35IEE626 pKa = 4.19FVDD629 pKa = 3.78NSKK632 pKa = 10.39VCYY635 pKa = 10.11ISMKK639 pKa = 10.03GRR641 pKa = 11.84NDD643 pKa = 3.62TIKK646 pKa = 10.7IMFNTTYY653 pKa = 9.91LGSMDD658 pKa = 4.08PSKK661 pKa = 10.94SIYY664 pKa = 9.03ATSSRR669 pKa = 11.84MDD671 pKa = 5.38DD672 pKa = 2.83ITGLVTSSGFINGIISTIGSISLFTIIGTFATRR705 pKa = 11.84LIGVILTMRR714 pKa = 11.84SVDD717 pKa = 3.46KK718 pKa = 10.87EE719 pKa = 4.42SYY721 pKa = 9.92RR722 pKa = 11.84SNAIII727 pKa = 4.38
Molecular weight: 81.65 kDa
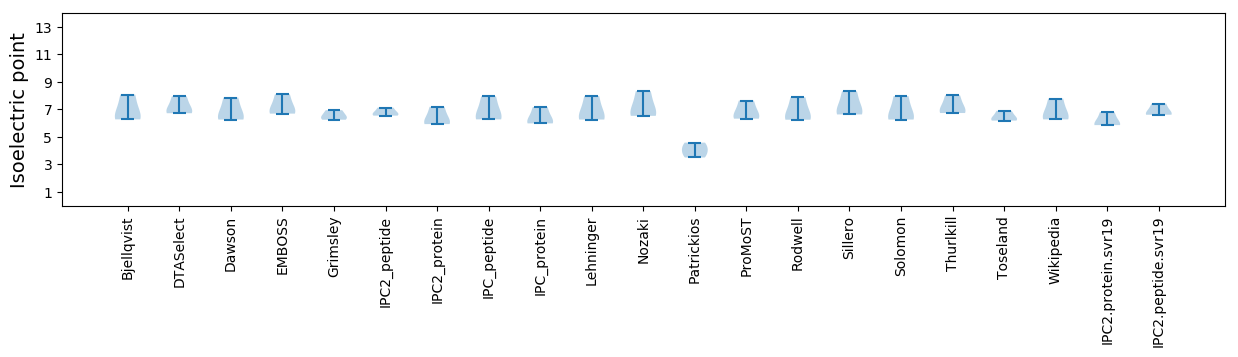
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A059XT43|A0A059XT43_9VIRU Nucleoprotein OS=Kigluaik phantom orthophasmavirus OX=1980539 GN=N PE=4 SV=1
MM1 pKa = 8.16DD2 pKa = 4.9KK3 pKa = 10.51PSEE6 pKa = 4.16AFTTAEE12 pKa = 3.58NSMVGLNLEE21 pKa = 4.54GPVQEE26 pKa = 4.67AVRR29 pKa = 11.84KK30 pKa = 7.5TFDD33 pKa = 3.16MHH35 pKa = 6.89AKK37 pKa = 9.55TIATMASHH45 pKa = 7.09PCSRR49 pKa = 11.84DD50 pKa = 3.02YY51 pKa = 12.1NMFLNTCEE59 pKa = 4.45SVSKK63 pKa = 9.98EE64 pKa = 3.98YY65 pKa = 11.25NSAGLDD71 pKa = 3.49AEE73 pKa = 4.39KK74 pKa = 10.62LCRR77 pKa = 11.84EE78 pKa = 3.97MSYY81 pKa = 11.33VNPDD85 pKa = 3.09FMLEE89 pKa = 4.1PGWEE93 pKa = 4.21VGHH96 pKa = 6.22QALNFMAKK104 pKa = 10.12VIFEE108 pKa = 4.39VGPEE112 pKa = 3.88TRR114 pKa = 11.84SVKK117 pKa = 10.53VGSGTDD123 pKa = 2.78KK124 pKa = 10.42SYY126 pKa = 11.04RR127 pKa = 11.84FRR129 pKa = 11.84FVRR132 pKa = 11.84QYY134 pKa = 11.23DD135 pKa = 3.29AADD138 pKa = 3.31IGRR141 pKa = 11.84VASKK145 pKa = 9.68MGAKK149 pKa = 9.89NPPNAPSTSAARR161 pKa = 11.84PNAVDD166 pKa = 3.45VVTAYY171 pKa = 9.63KK172 pKa = 10.68AKK174 pKa = 9.91IRR176 pKa = 11.84EE177 pKa = 4.03HH178 pKa = 7.17SVEE181 pKa = 3.97NGEE184 pKa = 4.18AGTVVSGMVIVKK196 pKa = 8.77TYY198 pKa = 10.61KK199 pKa = 9.47NTIAPQSEE207 pKa = 4.11PTVKK211 pKa = 10.68NGVLVLTIKK220 pKa = 10.31QASFLALYY228 pKa = 9.9KK229 pKa = 10.49FKK231 pKa = 11.01EE232 pKa = 4.22FVKK235 pKa = 10.6VCVARR240 pKa = 11.84DD241 pKa = 3.77TTILTPLCGAIFSRR255 pKa = 11.84DD256 pKa = 4.07DD257 pKa = 3.59IDD259 pKa = 6.19LMLADD264 pKa = 4.83PEE266 pKa = 4.21IKK268 pKa = 10.45AVITDD273 pKa = 3.56KK274 pKa = 11.48TEE276 pKa = 4.04LLVALNLSAQNGGQFVEE293 pKa = 5.18GSRR296 pKa = 11.84TDD298 pKa = 3.12IAGVCAIMGTVNVEE312 pKa = 3.78KK313 pKa = 10.4KK314 pKa = 10.67LRR316 pKa = 11.84MQIISKK322 pKa = 7.34TAKK325 pKa = 9.81QFLAKK330 pKa = 10.07DD331 pKa = 3.39RR332 pKa = 11.84PMDD335 pKa = 3.76KK336 pKa = 11.03GLFHH340 pKa = 7.49AVGKK344 pKa = 9.64YY345 pKa = 9.8AAGGLPAEE353 pKa = 4.36WQAEE357 pKa = 4.05ILVQEE362 pKa = 4.59LEE364 pKa = 4.18AAKK367 pKa = 8.82ITYY370 pKa = 9.63RR371 pKa = 11.84QILAAKK377 pKa = 9.28KK378 pKa = 8.07ATEE381 pKa = 3.64RR382 pKa = 11.84AIRR385 pKa = 11.84MEE387 pKa = 4.12VPNFDD392 pKa = 3.4GSEE395 pKa = 3.82NN396 pKa = 3.52
MM1 pKa = 8.16DD2 pKa = 4.9KK3 pKa = 10.51PSEE6 pKa = 4.16AFTTAEE12 pKa = 3.58NSMVGLNLEE21 pKa = 4.54GPVQEE26 pKa = 4.67AVRR29 pKa = 11.84KK30 pKa = 7.5TFDD33 pKa = 3.16MHH35 pKa = 6.89AKK37 pKa = 9.55TIATMASHH45 pKa = 7.09PCSRR49 pKa = 11.84DD50 pKa = 3.02YY51 pKa = 12.1NMFLNTCEE59 pKa = 4.45SVSKK63 pKa = 9.98EE64 pKa = 3.98YY65 pKa = 11.25NSAGLDD71 pKa = 3.49AEE73 pKa = 4.39KK74 pKa = 10.62LCRR77 pKa = 11.84EE78 pKa = 3.97MSYY81 pKa = 11.33VNPDD85 pKa = 3.09FMLEE89 pKa = 4.1PGWEE93 pKa = 4.21VGHH96 pKa = 6.22QALNFMAKK104 pKa = 10.12VIFEE108 pKa = 4.39VGPEE112 pKa = 3.88TRR114 pKa = 11.84SVKK117 pKa = 10.53VGSGTDD123 pKa = 2.78KK124 pKa = 10.42SYY126 pKa = 11.04RR127 pKa = 11.84FRR129 pKa = 11.84FVRR132 pKa = 11.84QYY134 pKa = 11.23DD135 pKa = 3.29AADD138 pKa = 3.31IGRR141 pKa = 11.84VASKK145 pKa = 9.68MGAKK149 pKa = 9.89NPPNAPSTSAARR161 pKa = 11.84PNAVDD166 pKa = 3.45VVTAYY171 pKa = 9.63KK172 pKa = 10.68AKK174 pKa = 9.91IRR176 pKa = 11.84EE177 pKa = 4.03HH178 pKa = 7.17SVEE181 pKa = 3.97NGEE184 pKa = 4.18AGTVVSGMVIVKK196 pKa = 8.77TYY198 pKa = 10.61KK199 pKa = 9.47NTIAPQSEE207 pKa = 4.11PTVKK211 pKa = 10.68NGVLVLTIKK220 pKa = 10.31QASFLALYY228 pKa = 9.9KK229 pKa = 10.49FKK231 pKa = 11.01EE232 pKa = 4.22FVKK235 pKa = 10.6VCVARR240 pKa = 11.84DD241 pKa = 3.77TTILTPLCGAIFSRR255 pKa = 11.84DD256 pKa = 4.07DD257 pKa = 3.59IDD259 pKa = 6.19LMLADD264 pKa = 4.83PEE266 pKa = 4.21IKK268 pKa = 10.45AVITDD273 pKa = 3.56KK274 pKa = 11.48TEE276 pKa = 4.04LLVALNLSAQNGGQFVEE293 pKa = 5.18GSRR296 pKa = 11.84TDD298 pKa = 3.12IAGVCAIMGTVNVEE312 pKa = 3.78KK313 pKa = 10.4KK314 pKa = 10.67LRR316 pKa = 11.84MQIISKK322 pKa = 7.34TAKK325 pKa = 9.81QFLAKK330 pKa = 10.07DD331 pKa = 3.39RR332 pKa = 11.84PMDD335 pKa = 3.76KK336 pKa = 11.03GLFHH340 pKa = 7.49AVGKK344 pKa = 9.64YY345 pKa = 9.8AAGGLPAEE353 pKa = 4.36WQAEE357 pKa = 4.05ILVQEE362 pKa = 4.59LEE364 pKa = 4.18AAKK367 pKa = 8.82ITYY370 pKa = 9.63RR371 pKa = 11.84QILAAKK377 pKa = 9.28KK378 pKa = 8.07ATEE381 pKa = 3.64RR382 pKa = 11.84AIRR385 pKa = 11.84MEE387 pKa = 4.12VPNFDD392 pKa = 3.4GSEE395 pKa = 3.82NN396 pKa = 3.52
Molecular weight: 43.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3254 |
396 |
2131 |
1084.7 |
123.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.948 ± 1.552 | 1.967 ± 0.616 |
6.146 ± 0.522 | 6.361 ± 0.473 |
3.749 ± 0.072 | 5.47 ± 0.589 |
1.936 ± 0.249 | 8.543 ± 1.686 |
6.976 ± 0.306 | 7.468 ± 0.926 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.473 ± 0.153 | 7.037 ± 0.686 |
3.258 ± 0.361 | 2.612 ± 0.415 |
4.948 ± 0.445 | 7.99 ± 1.071 |
5.655 ± 0.224 | 5.747 ± 0.745 |
0.707 ± 0.204 | 5.009 ± 0.665 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
