
Maricaulis salignorans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Maricaulales; Maricaulaceae; Maricaulis
Average proteome isoelectric point is 5.97
Get precalculated fractions of proteins
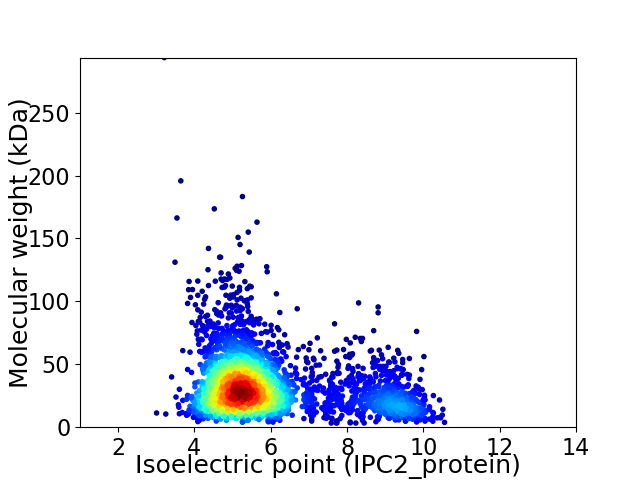
Virtual 2D-PAGE plot for 2949 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G9LDN2|A0A1G9LDN2_9PROT Uncharacterized protein OS=Maricaulis salignorans OX=144026 GN=SAMN04488568_1014 PE=4 SV=1
MM1 pKa = 7.89ASTLISGLAVAAPAMAQDD19 pKa = 3.61QDD21 pKa = 4.06VVTVTGSRR29 pKa = 11.84IQRR32 pKa = 11.84TDD34 pKa = 3.36TIAPSPVTNVSAEE47 pKa = 3.82QLAVVNTVNTEE58 pKa = 3.94DD59 pKa = 5.14FINTLPQVVPAFDD72 pKa = 3.39ATSNNPGDD80 pKa = 3.87GTATVALRR88 pKa = 11.84GLGATRR94 pKa = 11.84TLVLVDD100 pKa = 3.33GQRR103 pKa = 11.84FVGSGVSQVVDD114 pKa = 4.28LNNIPAAMVEE124 pKa = 4.45SIDD127 pKa = 3.68IVTGGASAVYY137 pKa = 10.33GSDD140 pKa = 2.82AVAGVVNFMMKK151 pKa = 10.48DD152 pKa = 3.18DD153 pKa = 4.13FEE155 pKa = 4.8GVEE158 pKa = 3.97ITASQEE164 pKa = 3.83TSLGDD169 pKa = 3.72LDD171 pKa = 5.79ADD173 pKa = 3.59ITNVSITMGGNFDD186 pKa = 3.78NGRR189 pKa = 11.84GNAVLSMGYY198 pKa = 7.5TNRR201 pKa = 11.84QALFQGDD208 pKa = 3.8RR209 pKa = 11.84AFSALTNWDD218 pKa = 3.84PGPGNEE224 pKa = 4.87AAGFYY229 pKa = 10.54AGGSSNIPGTRR240 pKa = 11.84IRR242 pKa = 11.84GGGSTNFGLMNYY254 pKa = 8.7DD255 pKa = 3.41ANGNEE260 pKa = 4.08IDD262 pKa = 4.39SSNTSYY268 pKa = 10.8LASDD272 pKa = 4.74RR273 pKa = 11.84IYY275 pKa = 10.71DD276 pKa = 3.94IFPEE280 pKa = 4.71CGAGNALACGGVFNDD295 pKa = 3.77GGIRR299 pKa = 11.84GMRR302 pKa = 11.84FGGATPDD309 pKa = 3.42VYY311 pKa = 11.41NYY313 pKa = 11.18APANYY318 pKa = 9.33LQLPQEE324 pKa = 4.68RR325 pKa = 11.84YY326 pKa = 9.96NISAFATYY334 pKa = 9.5EE335 pKa = 4.19VNEE338 pKa = 3.96HH339 pKa = 6.34LEE341 pKa = 4.21FYY343 pKa = 11.16GRR345 pKa = 11.84GIYY348 pKa = 10.3ANNVVDD354 pKa = 4.2SQLAPTPAGVTLNVNLDD371 pKa = 3.9NPNLPADD378 pKa = 4.09LVDD381 pKa = 4.86LLVGDD386 pKa = 4.14VDD388 pKa = 4.23AVVATGLADD397 pKa = 4.57DD398 pKa = 5.52PGTAADD404 pKa = 3.54EE405 pKa = 4.33SVYY408 pKa = 11.1NHH410 pKa = 6.46SGSNNGDD417 pKa = 2.89GTATVRR423 pKa = 11.84ISRR426 pKa = 11.84RR427 pKa = 11.84FEE429 pKa = 4.25EE430 pKa = 3.87IGTRR434 pKa = 11.84NSLRR438 pKa = 11.84DD439 pKa = 3.55TNSFQVVFGARR450 pKa = 11.84GALNEE455 pKa = 4.02NWSYY459 pKa = 11.86DD460 pKa = 2.89AFANYY465 pKa = 9.45SRR467 pKa = 11.84SAVSQIQTGNLSVSALQDD485 pKa = 3.31GVLCDD490 pKa = 4.28GGPTALASGCSAPYY504 pKa = 10.26VDD506 pKa = 5.52LFNGPGAISAAGASHH521 pKa = 7.62ISRR524 pKa = 11.84TGAQIDD530 pKa = 4.32TIEE533 pKa = 4.14MFQTQATVNGVLPMFQSPAANEE555 pKa = 3.78PLALVLGVEE564 pKa = 4.23YY565 pKa = 10.8RR566 pKa = 11.84EE567 pKa = 4.36ASSNSLPDD575 pKa = 3.51SVLGPDD581 pKa = 3.33VRR583 pKa = 11.84GFNQALPVGGTVDD596 pKa = 3.12VYY598 pKa = 11.48EE599 pKa = 4.66FFTEE603 pKa = 4.26VEE605 pKa = 4.15MPLIEE610 pKa = 5.12DD611 pKa = 3.55RR612 pKa = 11.84PGIQSLSLNAAFRR625 pKa = 11.84SSEE628 pKa = 4.01YY629 pKa = 9.21NTVGGTEE636 pKa = 4.04TFAVGLGWEE645 pKa = 4.38VNDD648 pKa = 4.59QIRR651 pKa = 11.84LRR653 pKa = 11.84GNFNRR658 pKa = 11.84AVRR661 pKa = 11.84APNVGDD667 pKa = 4.35LYY669 pKa = 11.18QPLVNGFPSAKK680 pKa = 9.58DD681 pKa = 3.38PCSGGSNGSYY691 pKa = 10.39GAATIDD697 pKa = 3.65QTCVDD702 pKa = 4.65AGVPAPGAAIQGNAQMEE719 pKa = 4.44ALFGGNPNLDD729 pKa = 3.59AEE731 pKa = 4.6TADD734 pKa = 3.65TYY736 pKa = 11.12TIGVVLTPDD745 pKa = 4.16AIEE748 pKa = 4.47GLTMTLDD755 pKa = 3.69YY756 pKa = 11.24YY757 pKa = 11.28NISIEE762 pKa = 4.22DD763 pKa = 4.56AISTVPLQTLLNEE776 pKa = 4.09CHH778 pKa = 6.93IDD780 pKa = 4.31NIAASCTALAGGRR793 pKa = 11.84GPSGEE798 pKa = 3.93MGANGFLPSLVSINVASLEE817 pKa = 4.05AEE819 pKa = 4.78GIDD822 pKa = 4.31LRR824 pKa = 11.84VDD826 pKa = 3.29YY827 pKa = 11.31GFGADD832 pKa = 3.7VLGLPGDD839 pKa = 4.02FAVTYY844 pKa = 9.98YY845 pKa = 10.94GGYY848 pKa = 8.82TIANNFTPSASSPVVEE864 pKa = 4.36CAGFFGLDD872 pKa = 3.54CGEE875 pKa = 4.04PTPEE879 pKa = 4.54YY880 pKa = 9.19KK881 pKa = 10.4HH882 pKa = 6.85SMQASWYY889 pKa = 7.53TGPFTTSLRR898 pKa = 11.84WRR900 pKa = 11.84YY901 pKa = 9.61IGAVEE906 pKa = 4.57ADD908 pKa = 3.83SATAPQVSSLSDD920 pKa = 3.69SIDD923 pKa = 3.01ATNYY927 pKa = 10.05FDD929 pKa = 4.64VTFQYY934 pKa = 10.8DD935 pKa = 3.28INDD938 pKa = 3.96TFTLTTGISNITDD951 pKa = 3.6TEE953 pKa = 4.55VPAIGSTSTEE963 pKa = 4.13QANTWPATYY972 pKa = 7.55EE973 pKa = 3.96TLGRR977 pKa = 11.84RR978 pKa = 11.84LFVGGTMRR986 pKa = 11.84FF987 pKa = 3.5
MM1 pKa = 7.89ASTLISGLAVAAPAMAQDD19 pKa = 3.61QDD21 pKa = 4.06VVTVTGSRR29 pKa = 11.84IQRR32 pKa = 11.84TDD34 pKa = 3.36TIAPSPVTNVSAEE47 pKa = 3.82QLAVVNTVNTEE58 pKa = 3.94DD59 pKa = 5.14FINTLPQVVPAFDD72 pKa = 3.39ATSNNPGDD80 pKa = 3.87GTATVALRR88 pKa = 11.84GLGATRR94 pKa = 11.84TLVLVDD100 pKa = 3.33GQRR103 pKa = 11.84FVGSGVSQVVDD114 pKa = 4.28LNNIPAAMVEE124 pKa = 4.45SIDD127 pKa = 3.68IVTGGASAVYY137 pKa = 10.33GSDD140 pKa = 2.82AVAGVVNFMMKK151 pKa = 10.48DD152 pKa = 3.18DD153 pKa = 4.13FEE155 pKa = 4.8GVEE158 pKa = 3.97ITASQEE164 pKa = 3.83TSLGDD169 pKa = 3.72LDD171 pKa = 5.79ADD173 pKa = 3.59ITNVSITMGGNFDD186 pKa = 3.78NGRR189 pKa = 11.84GNAVLSMGYY198 pKa = 7.5TNRR201 pKa = 11.84QALFQGDD208 pKa = 3.8RR209 pKa = 11.84AFSALTNWDD218 pKa = 3.84PGPGNEE224 pKa = 4.87AAGFYY229 pKa = 10.54AGGSSNIPGTRR240 pKa = 11.84IRR242 pKa = 11.84GGGSTNFGLMNYY254 pKa = 8.7DD255 pKa = 3.41ANGNEE260 pKa = 4.08IDD262 pKa = 4.39SSNTSYY268 pKa = 10.8LASDD272 pKa = 4.74RR273 pKa = 11.84IYY275 pKa = 10.71DD276 pKa = 3.94IFPEE280 pKa = 4.71CGAGNALACGGVFNDD295 pKa = 3.77GGIRR299 pKa = 11.84GMRR302 pKa = 11.84FGGATPDD309 pKa = 3.42VYY311 pKa = 11.41NYY313 pKa = 11.18APANYY318 pKa = 9.33LQLPQEE324 pKa = 4.68RR325 pKa = 11.84YY326 pKa = 9.96NISAFATYY334 pKa = 9.5EE335 pKa = 4.19VNEE338 pKa = 3.96HH339 pKa = 6.34LEE341 pKa = 4.21FYY343 pKa = 11.16GRR345 pKa = 11.84GIYY348 pKa = 10.3ANNVVDD354 pKa = 4.2SQLAPTPAGVTLNVNLDD371 pKa = 3.9NPNLPADD378 pKa = 4.09LVDD381 pKa = 4.86LLVGDD386 pKa = 4.14VDD388 pKa = 4.23AVVATGLADD397 pKa = 4.57DD398 pKa = 5.52PGTAADD404 pKa = 3.54EE405 pKa = 4.33SVYY408 pKa = 11.1NHH410 pKa = 6.46SGSNNGDD417 pKa = 2.89GTATVRR423 pKa = 11.84ISRR426 pKa = 11.84RR427 pKa = 11.84FEE429 pKa = 4.25EE430 pKa = 3.87IGTRR434 pKa = 11.84NSLRR438 pKa = 11.84DD439 pKa = 3.55TNSFQVVFGARR450 pKa = 11.84GALNEE455 pKa = 4.02NWSYY459 pKa = 11.86DD460 pKa = 2.89AFANYY465 pKa = 9.45SRR467 pKa = 11.84SAVSQIQTGNLSVSALQDD485 pKa = 3.31GVLCDD490 pKa = 4.28GGPTALASGCSAPYY504 pKa = 10.26VDD506 pKa = 5.52LFNGPGAISAAGASHH521 pKa = 7.62ISRR524 pKa = 11.84TGAQIDD530 pKa = 4.32TIEE533 pKa = 4.14MFQTQATVNGVLPMFQSPAANEE555 pKa = 3.78PLALVLGVEE564 pKa = 4.23YY565 pKa = 10.8RR566 pKa = 11.84EE567 pKa = 4.36ASSNSLPDD575 pKa = 3.51SVLGPDD581 pKa = 3.33VRR583 pKa = 11.84GFNQALPVGGTVDD596 pKa = 3.12VYY598 pKa = 11.48EE599 pKa = 4.66FFTEE603 pKa = 4.26VEE605 pKa = 4.15MPLIEE610 pKa = 5.12DD611 pKa = 3.55RR612 pKa = 11.84PGIQSLSLNAAFRR625 pKa = 11.84SSEE628 pKa = 4.01YY629 pKa = 9.21NTVGGTEE636 pKa = 4.04TFAVGLGWEE645 pKa = 4.38VNDD648 pKa = 4.59QIRR651 pKa = 11.84LRR653 pKa = 11.84GNFNRR658 pKa = 11.84AVRR661 pKa = 11.84APNVGDD667 pKa = 4.35LYY669 pKa = 11.18QPLVNGFPSAKK680 pKa = 9.58DD681 pKa = 3.38PCSGGSNGSYY691 pKa = 10.39GAATIDD697 pKa = 3.65QTCVDD702 pKa = 4.65AGVPAPGAAIQGNAQMEE719 pKa = 4.44ALFGGNPNLDD729 pKa = 3.59AEE731 pKa = 4.6TADD734 pKa = 3.65TYY736 pKa = 11.12TIGVVLTPDD745 pKa = 4.16AIEE748 pKa = 4.47GLTMTLDD755 pKa = 3.69YY756 pKa = 11.24YY757 pKa = 11.28NISIEE762 pKa = 4.22DD763 pKa = 4.56AISTVPLQTLLNEE776 pKa = 4.09CHH778 pKa = 6.93IDD780 pKa = 4.31NIAASCTALAGGRR793 pKa = 11.84GPSGEE798 pKa = 3.93MGANGFLPSLVSINVASLEE817 pKa = 4.05AEE819 pKa = 4.78GIDD822 pKa = 4.31LRR824 pKa = 11.84VDD826 pKa = 3.29YY827 pKa = 11.31GFGADD832 pKa = 3.7VLGLPGDD839 pKa = 4.02FAVTYY844 pKa = 9.98YY845 pKa = 10.94GGYY848 pKa = 8.82TIANNFTPSASSPVVEE864 pKa = 4.36CAGFFGLDD872 pKa = 3.54CGEE875 pKa = 4.04PTPEE879 pKa = 4.54YY880 pKa = 9.19KK881 pKa = 10.4HH882 pKa = 6.85SMQASWYY889 pKa = 7.53TGPFTTSLRR898 pKa = 11.84WRR900 pKa = 11.84YY901 pKa = 9.61IGAVEE906 pKa = 4.57ADD908 pKa = 3.83SATAPQVSSLSDD920 pKa = 3.69SIDD923 pKa = 3.01ATNYY927 pKa = 10.05FDD929 pKa = 4.64VTFQYY934 pKa = 10.8DD935 pKa = 3.28INDD938 pKa = 3.96TFTLTTGISNITDD951 pKa = 3.6TEE953 pKa = 4.55VPAIGSTSTEE963 pKa = 4.13QANTWPATYY972 pKa = 7.55EE973 pKa = 3.96TLGRR977 pKa = 11.84RR978 pKa = 11.84LFVGGTMRR986 pKa = 11.84FF987 pKa = 3.5
Molecular weight: 103.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G9SQW3|A0A1G9SQW3_9PROT Lipopolysaccharide export system ATP-binding protein LptB OS=Maricaulis salignorans OX=144026 GN=SAMN04488568_11042 PE=3 SV=1
MM1 pKa = 6.89TQTGRR6 pKa = 11.84IRR8 pKa = 11.84LGDD11 pKa = 3.39KK12 pKa = 10.29DD13 pKa = 3.09IPYY16 pKa = 10.86ALRR19 pKa = 11.84RR20 pKa = 11.84STRR23 pKa = 11.84RR24 pKa = 11.84TIGFVINRR32 pKa = 11.84DD33 pKa = 3.75GLSVTAPQRR42 pKa = 11.84VSVRR46 pKa = 11.84DD47 pKa = 3.37IEE49 pKa = 4.7AALQEE54 pKa = 4.19KK55 pKa = 8.72SHH57 pKa = 6.7WILGKK62 pKa = 9.89LAAWAEE68 pKa = 4.21RR69 pKa = 11.84PHH71 pKa = 6.51QRR73 pKa = 11.84EE74 pKa = 3.72LAYY77 pKa = 9.82VTGEE81 pKa = 4.0TLPWLGGEE89 pKa = 4.68LKK91 pKa = 10.83LQVEE95 pKa = 4.52PRR97 pKa = 11.84GIRR100 pKa = 11.84TTLRR104 pKa = 11.84RR105 pKa = 11.84DD106 pKa = 3.09GDD108 pKa = 3.9RR109 pKa = 11.84VLVTVDD115 pKa = 3.88PEE117 pKa = 4.01ASGEE121 pKa = 3.94LRR123 pKa = 11.84RR124 pKa = 11.84RR125 pKa = 11.84AIASALMRR133 pKa = 11.84HH134 pKa = 5.67YY135 pKa = 10.47KK136 pKa = 10.27RR137 pKa = 11.84EE138 pKa = 4.17GEE140 pKa = 4.02HH141 pKa = 5.98FMRR144 pKa = 11.84PRR146 pKa = 11.84VEE148 pKa = 4.72AFATRR153 pKa = 11.84LEE155 pKa = 4.16RR156 pKa = 11.84PVRR159 pKa = 11.84KK160 pKa = 9.7VVIRR164 pKa = 11.84DD165 pKa = 3.28QKK167 pKa = 10.82RR168 pKa = 11.84RR169 pKa = 11.84WGSCAPDD176 pKa = 3.4GTIRR180 pKa = 11.84LNWRR184 pKa = 11.84LMGFPEE190 pKa = 3.94NLVDD194 pKa = 3.84YY195 pKa = 10.59VCAHH199 pKa = 6.72EE200 pKa = 4.47AAHH203 pKa = 6.27LVEE206 pKa = 5.54HH207 pKa = 6.0NHH209 pKa = 6.19SPAFWKK215 pKa = 9.05TVEE218 pKa = 4.95RR219 pKa = 11.84IMPDD223 pKa = 2.65WRR225 pKa = 11.84IHH227 pKa = 5.69RR228 pKa = 11.84DD229 pKa = 3.31EE230 pKa = 4.19MRR232 pKa = 11.84RR233 pKa = 11.84AADD236 pKa = 3.05LWTWVV241 pKa = 3.38
MM1 pKa = 6.89TQTGRR6 pKa = 11.84IRR8 pKa = 11.84LGDD11 pKa = 3.39KK12 pKa = 10.29DD13 pKa = 3.09IPYY16 pKa = 10.86ALRR19 pKa = 11.84RR20 pKa = 11.84STRR23 pKa = 11.84RR24 pKa = 11.84TIGFVINRR32 pKa = 11.84DD33 pKa = 3.75GLSVTAPQRR42 pKa = 11.84VSVRR46 pKa = 11.84DD47 pKa = 3.37IEE49 pKa = 4.7AALQEE54 pKa = 4.19KK55 pKa = 8.72SHH57 pKa = 6.7WILGKK62 pKa = 9.89LAAWAEE68 pKa = 4.21RR69 pKa = 11.84PHH71 pKa = 6.51QRR73 pKa = 11.84EE74 pKa = 3.72LAYY77 pKa = 9.82VTGEE81 pKa = 4.0TLPWLGGEE89 pKa = 4.68LKK91 pKa = 10.83LQVEE95 pKa = 4.52PRR97 pKa = 11.84GIRR100 pKa = 11.84TTLRR104 pKa = 11.84RR105 pKa = 11.84DD106 pKa = 3.09GDD108 pKa = 3.9RR109 pKa = 11.84VLVTVDD115 pKa = 3.88PEE117 pKa = 4.01ASGEE121 pKa = 3.94LRR123 pKa = 11.84RR124 pKa = 11.84RR125 pKa = 11.84AIASALMRR133 pKa = 11.84HH134 pKa = 5.67YY135 pKa = 10.47KK136 pKa = 10.27RR137 pKa = 11.84EE138 pKa = 4.17GEE140 pKa = 4.02HH141 pKa = 5.98FMRR144 pKa = 11.84PRR146 pKa = 11.84VEE148 pKa = 4.72AFATRR153 pKa = 11.84LEE155 pKa = 4.16RR156 pKa = 11.84PVRR159 pKa = 11.84KK160 pKa = 9.7VVIRR164 pKa = 11.84DD165 pKa = 3.28QKK167 pKa = 10.82RR168 pKa = 11.84RR169 pKa = 11.84WGSCAPDD176 pKa = 3.4GTIRR180 pKa = 11.84LNWRR184 pKa = 11.84LMGFPEE190 pKa = 3.94NLVDD194 pKa = 3.84YY195 pKa = 10.59VCAHH199 pKa = 6.72EE200 pKa = 4.47AAHH203 pKa = 6.27LVEE206 pKa = 5.54HH207 pKa = 6.0NHH209 pKa = 6.19SPAFWKK215 pKa = 9.05TVEE218 pKa = 4.95RR219 pKa = 11.84IMPDD223 pKa = 2.65WRR225 pKa = 11.84IHH227 pKa = 5.69RR228 pKa = 11.84DD229 pKa = 3.31EE230 pKa = 4.19MRR232 pKa = 11.84RR233 pKa = 11.84AADD236 pKa = 3.05LWTWVV241 pKa = 3.38
Molecular weight: 28.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
939932 |
29 |
2981 |
318.7 |
34.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.041 ± 0.063 | 0.825 ± 0.013 |
6.226 ± 0.041 | 5.948 ± 0.041 |
3.682 ± 0.026 | 8.726 ± 0.049 |
1.974 ± 0.02 | 5.243 ± 0.031 |
2.49 ± 0.043 | 10.058 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.522 ± 0.022 | 2.557 ± 0.024 |
5.022 ± 0.033 | 3.174 ± 0.021 |
7.173 ± 0.053 | 5.565 ± 0.037 |
5.248 ± 0.037 | 6.906 ± 0.037 |
1.408 ± 0.02 | 2.213 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
