
Black robin associated gemykibivirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykibivirus; Gemykibivirus blaro1
Average proteome isoelectric point is 7.36
Get precalculated fractions of proteins
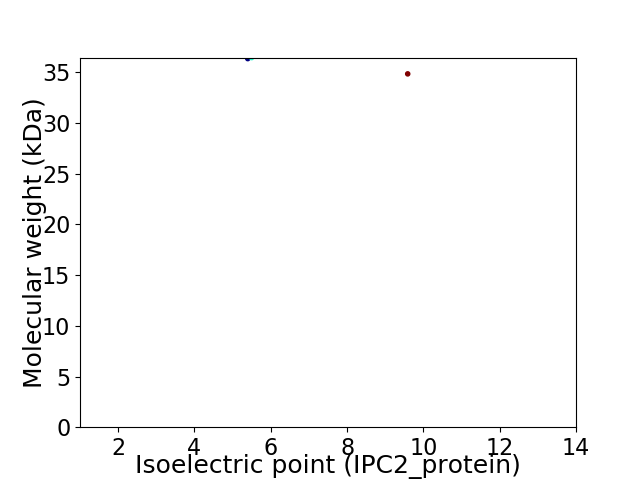
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|T1YTB1|T1YTB1_9VIRU Capsid protein OS=Black robin associated gemykibivirus 1 OX=1391037 PE=4 SV=1
MM1 pKa = 7.58SFRR4 pKa = 11.84FAAKK8 pKa = 9.17WGLLTFPQSGDD19 pKa = 3.37LDD21 pKa = 3.46PWEE24 pKa = 4.67VNDD27 pKa = 3.89HH28 pKa = 6.64LGRR31 pKa = 11.84LGAEE35 pKa = 4.19CIIGRR40 pKa = 11.84EE41 pKa = 3.94EE42 pKa = 3.89HH43 pKa = 7.07ADD45 pKa = 3.59GGLHH49 pKa = 5.95LHH51 pKa = 6.48AFFMFEE57 pKa = 4.26RR58 pKa = 11.84KK59 pKa = 9.22FEE61 pKa = 4.24SRR63 pKa = 11.84NVRR66 pKa = 11.84VFDD69 pKa = 3.45VDD71 pKa = 3.28GRR73 pKa = 11.84HH74 pKa = 5.89PNVVRR79 pKa = 11.84GYY81 pKa = 7.82STPEE85 pKa = 3.33KK86 pKa = 10.04GATYY90 pKa = 10.03AIKK93 pKa = 10.89DD94 pKa = 3.57GDD96 pKa = 4.14VVAGGLDD103 pKa = 3.73PASLGGDD110 pKa = 2.96RR111 pKa = 11.84VDD113 pKa = 4.75RR114 pKa = 11.84VADD117 pKa = 3.95PWPAIILSTTRR128 pKa = 11.84DD129 pKa = 3.6EE130 pKa = 4.86FFEE133 pKa = 4.4ACARR137 pKa = 11.84MAPKK141 pKa = 10.52SLVCSFNSLKK151 pKa = 10.72CYY153 pKa = 10.53ADD155 pKa = 2.72WRR157 pKa = 11.84YY158 pKa = 9.95RR159 pKa = 11.84PEE161 pKa = 4.12PVPYY165 pKa = 8.89EE166 pKa = 4.25HH167 pKa = 7.51PSGVQICTDD176 pKa = 4.15RR177 pKa = 11.84FPDD180 pKa = 3.66LSSWVLQNLEE190 pKa = 4.03RR191 pKa = 11.84RR192 pKa = 11.84TSGRR196 pKa = 11.84RR197 pKa = 11.84RR198 pKa = 11.84SLVLYY203 pKa = 10.58GPTRR207 pKa = 11.84LGKK210 pKa = 7.5TLWARR215 pKa = 11.84SLGNHH220 pKa = 7.12AYY222 pKa = 10.41FGGLFSLEE230 pKa = 4.18EE231 pKa = 3.98EE232 pKa = 4.7LEE234 pKa = 4.33GVDD237 pKa = 4.11YY238 pKa = 11.31AIFDD242 pKa = 4.11DD243 pKa = 4.11MQGGLKK249 pKa = 10.09FFHH252 pKa = 7.11SYY254 pKa = 10.7KK255 pKa = 10.25FWLGAQSQFWATDD268 pKa = 2.95KK269 pKa = 11.37YY270 pKa = 10.54RR271 pKa = 11.84GKK273 pKa = 10.83KK274 pKa = 8.9LIHH277 pKa = 6.62WGRR280 pKa = 11.84PSIYY284 pKa = 9.68ICNQNPLCDD293 pKa = 3.74EE294 pKa = 4.6GVDD297 pKa = 3.91HH298 pKa = 7.77DD299 pKa = 4.55WLIGNCDD306 pKa = 3.26FVEE309 pKa = 4.24ITEE312 pKa = 4.27SLLVPEE318 pKa = 5.05EE319 pKa = 4.32GG320 pKa = 3.94
MM1 pKa = 7.58SFRR4 pKa = 11.84FAAKK8 pKa = 9.17WGLLTFPQSGDD19 pKa = 3.37LDD21 pKa = 3.46PWEE24 pKa = 4.67VNDD27 pKa = 3.89HH28 pKa = 6.64LGRR31 pKa = 11.84LGAEE35 pKa = 4.19CIIGRR40 pKa = 11.84EE41 pKa = 3.94EE42 pKa = 3.89HH43 pKa = 7.07ADD45 pKa = 3.59GGLHH49 pKa = 5.95LHH51 pKa = 6.48AFFMFEE57 pKa = 4.26RR58 pKa = 11.84KK59 pKa = 9.22FEE61 pKa = 4.24SRR63 pKa = 11.84NVRR66 pKa = 11.84VFDD69 pKa = 3.45VDD71 pKa = 3.28GRR73 pKa = 11.84HH74 pKa = 5.89PNVVRR79 pKa = 11.84GYY81 pKa = 7.82STPEE85 pKa = 3.33KK86 pKa = 10.04GATYY90 pKa = 10.03AIKK93 pKa = 10.89DD94 pKa = 3.57GDD96 pKa = 4.14VVAGGLDD103 pKa = 3.73PASLGGDD110 pKa = 2.96RR111 pKa = 11.84VDD113 pKa = 4.75RR114 pKa = 11.84VADD117 pKa = 3.95PWPAIILSTTRR128 pKa = 11.84DD129 pKa = 3.6EE130 pKa = 4.86FFEE133 pKa = 4.4ACARR137 pKa = 11.84MAPKK141 pKa = 10.52SLVCSFNSLKK151 pKa = 10.72CYY153 pKa = 10.53ADD155 pKa = 2.72WRR157 pKa = 11.84YY158 pKa = 9.95RR159 pKa = 11.84PEE161 pKa = 4.12PVPYY165 pKa = 8.89EE166 pKa = 4.25HH167 pKa = 7.51PSGVQICTDD176 pKa = 4.15RR177 pKa = 11.84FPDD180 pKa = 3.66LSSWVLQNLEE190 pKa = 4.03RR191 pKa = 11.84RR192 pKa = 11.84TSGRR196 pKa = 11.84RR197 pKa = 11.84RR198 pKa = 11.84SLVLYY203 pKa = 10.58GPTRR207 pKa = 11.84LGKK210 pKa = 7.5TLWARR215 pKa = 11.84SLGNHH220 pKa = 7.12AYY222 pKa = 10.41FGGLFSLEE230 pKa = 4.18EE231 pKa = 3.98EE232 pKa = 4.7LEE234 pKa = 4.33GVDD237 pKa = 4.11YY238 pKa = 11.31AIFDD242 pKa = 4.11DD243 pKa = 4.11MQGGLKK249 pKa = 10.09FFHH252 pKa = 7.11SYY254 pKa = 10.7KK255 pKa = 10.25FWLGAQSQFWATDD268 pKa = 2.95KK269 pKa = 11.37YY270 pKa = 10.54RR271 pKa = 11.84GKK273 pKa = 10.83KK274 pKa = 8.9LIHH277 pKa = 6.62WGRR280 pKa = 11.84PSIYY284 pKa = 9.68ICNQNPLCDD293 pKa = 3.74EE294 pKa = 4.6GVDD297 pKa = 3.91HH298 pKa = 7.77DD299 pKa = 4.55WLIGNCDD306 pKa = 3.26FVEE309 pKa = 4.24ITEE312 pKa = 4.27SLLVPEE318 pKa = 5.05EE319 pKa = 4.32GG320 pKa = 3.94
Molecular weight: 36.36 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|T1YTB1|T1YTB1_9VIRU Capsid protein OS=Black robin associated gemykibivirus 1 OX=1391037 PE=4 SV=1
MM1 pKa = 7.72AYY3 pKa = 10.17ARR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84FTSSRR12 pKa = 11.84ARR14 pKa = 11.84KK15 pKa = 8.46PIRR18 pKa = 11.84RR19 pKa = 11.84SRR21 pKa = 11.84RR22 pKa = 11.84PAGKK26 pKa = 8.67RR27 pKa = 11.84RR28 pKa = 11.84SYY30 pKa = 10.65RR31 pKa = 11.84RR32 pKa = 11.84STRR35 pKa = 11.84KK36 pKa = 9.65RR37 pKa = 11.84GMSTRR42 pKa = 11.84TLLNKK47 pKa = 9.52TSQKK51 pKa = 10.41KK52 pKa = 9.12RR53 pKa = 11.84DD54 pKa = 3.61NMLSFSNTTAEE65 pKa = 4.83DD66 pKa = 4.11PFSTDD71 pKa = 3.1YY72 pKa = 11.17TIGPAVMRR80 pKa = 11.84RR81 pKa = 11.84PVGADD86 pKa = 3.39LPAEE90 pKa = 4.97FIYY93 pKa = 10.52VWNATGRR100 pKa = 11.84PHH102 pKa = 6.78EE103 pKa = 4.7LSTGFRR109 pKa = 11.84GSKK112 pKa = 9.67IDD114 pKa = 3.28TSLRR118 pKa = 11.84TNTTIFAVGLKK129 pKa = 10.01EE130 pKa = 4.46RR131 pKa = 11.84IKK133 pKa = 11.3LEE135 pKa = 4.44TNNSAPWRR143 pKa = 11.84WRR145 pKa = 11.84RR146 pKa = 11.84ICFTSKK152 pKa = 10.59NDD154 pKa = 3.53YY155 pKa = 11.27GEE157 pKa = 4.97ADD159 pKa = 4.21PDD161 pKa = 3.33EE162 pKa = 5.37SDD164 pKa = 3.53YY165 pKa = 11.01FRR167 pKa = 11.84RR168 pKa = 11.84TSNGMVRR175 pKa = 11.84LLRR178 pKa = 11.84AQSQAEE184 pKa = 4.46EE185 pKa = 4.27INDD188 pKa = 3.85DD189 pKa = 3.76VFDD192 pKa = 4.11GQRR195 pKa = 11.84NVDD198 pKa = 3.41WLSAITAPINRR209 pKa = 11.84RR210 pKa = 11.84HH211 pKa = 5.62VSVHH215 pKa = 5.13YY216 pKa = 10.69DD217 pKa = 3.19RR218 pKa = 11.84TRR220 pKa = 11.84TIQSGNQSGVTRR232 pKa = 11.84TYY234 pKa = 10.51RR235 pKa = 11.84QWHH238 pKa = 5.74PMRR241 pKa = 11.84KK242 pKa = 8.28NVVYY246 pKa = 10.61ADD248 pKa = 3.85EE249 pKa = 4.16QAGEE253 pKa = 4.32GMVDD257 pKa = 3.08SGVSVTGKK265 pKa = 10.38VGMGNYY271 pKa = 10.03YY272 pKa = 11.15VMDD275 pKa = 4.08IFTKK279 pKa = 10.52HH280 pKa = 5.92GLNDD284 pKa = 3.79DD285 pKa = 3.77QSTLSYY291 pKa = 10.27EE292 pKa = 4.2PEE294 pKa = 4.09STFFWHH300 pKa = 6.76EE301 pKa = 3.87KK302 pKa = 9.8
MM1 pKa = 7.72AYY3 pKa = 10.17ARR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84FTSSRR12 pKa = 11.84ARR14 pKa = 11.84KK15 pKa = 8.46PIRR18 pKa = 11.84RR19 pKa = 11.84SRR21 pKa = 11.84RR22 pKa = 11.84PAGKK26 pKa = 8.67RR27 pKa = 11.84RR28 pKa = 11.84SYY30 pKa = 10.65RR31 pKa = 11.84RR32 pKa = 11.84STRR35 pKa = 11.84KK36 pKa = 9.65RR37 pKa = 11.84GMSTRR42 pKa = 11.84TLLNKK47 pKa = 9.52TSQKK51 pKa = 10.41KK52 pKa = 9.12RR53 pKa = 11.84DD54 pKa = 3.61NMLSFSNTTAEE65 pKa = 4.83DD66 pKa = 4.11PFSTDD71 pKa = 3.1YY72 pKa = 11.17TIGPAVMRR80 pKa = 11.84RR81 pKa = 11.84PVGADD86 pKa = 3.39LPAEE90 pKa = 4.97FIYY93 pKa = 10.52VWNATGRR100 pKa = 11.84PHH102 pKa = 6.78EE103 pKa = 4.7LSTGFRR109 pKa = 11.84GSKK112 pKa = 9.67IDD114 pKa = 3.28TSLRR118 pKa = 11.84TNTTIFAVGLKK129 pKa = 10.01EE130 pKa = 4.46RR131 pKa = 11.84IKK133 pKa = 11.3LEE135 pKa = 4.44TNNSAPWRR143 pKa = 11.84WRR145 pKa = 11.84RR146 pKa = 11.84ICFTSKK152 pKa = 10.59NDD154 pKa = 3.53YY155 pKa = 11.27GEE157 pKa = 4.97ADD159 pKa = 4.21PDD161 pKa = 3.33EE162 pKa = 5.37SDD164 pKa = 3.53YY165 pKa = 11.01FRR167 pKa = 11.84RR168 pKa = 11.84TSNGMVRR175 pKa = 11.84LLRR178 pKa = 11.84AQSQAEE184 pKa = 4.46EE185 pKa = 4.27INDD188 pKa = 3.85DD189 pKa = 3.76VFDD192 pKa = 4.11GQRR195 pKa = 11.84NVDD198 pKa = 3.41WLSAITAPINRR209 pKa = 11.84RR210 pKa = 11.84HH211 pKa = 5.62VSVHH215 pKa = 5.13YY216 pKa = 10.69DD217 pKa = 3.19RR218 pKa = 11.84TRR220 pKa = 11.84TIQSGNQSGVTRR232 pKa = 11.84TYY234 pKa = 10.51RR235 pKa = 11.84QWHH238 pKa = 5.74PMRR241 pKa = 11.84KK242 pKa = 8.28NVVYY246 pKa = 10.61ADD248 pKa = 3.85EE249 pKa = 4.16QAGEE253 pKa = 4.32GMVDD257 pKa = 3.08SGVSVTGKK265 pKa = 10.38VGMGNYY271 pKa = 10.03YY272 pKa = 11.15VMDD275 pKa = 4.08IFTKK279 pKa = 10.52HH280 pKa = 5.92GLNDD284 pKa = 3.79DD285 pKa = 3.77QSTLSYY291 pKa = 10.27EE292 pKa = 4.2PEE294 pKa = 4.09STFFWHH300 pKa = 6.76EE301 pKa = 3.87KK302 pKa = 9.8
Molecular weight: 34.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
622 |
302 |
320 |
311.0 |
35.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.109 ± 0.098 | 1.447 ± 0.734 |
6.913 ± 0.409 | 5.627 ± 0.652 |
5.145 ± 0.771 | 8.199 ± 1.038 |
2.572 ± 0.385 | 3.859 ± 0.076 |
4.18 ± 0.3 | 6.913 ± 1.717 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.09 ± 0.586 | 4.019 ± 0.841 |
4.662 ± 0.453 | 2.572 ± 0.268 |
9.968 ± 1.721 | 7.556 ± 0.911 |
6.109 ± 1.863 | 5.788 ± 0.104 |
2.572 ± 0.385 | 3.698 ± 0.181 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
