
Hubei virga-like virus 23
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.92
Get precalculated fractions of proteins
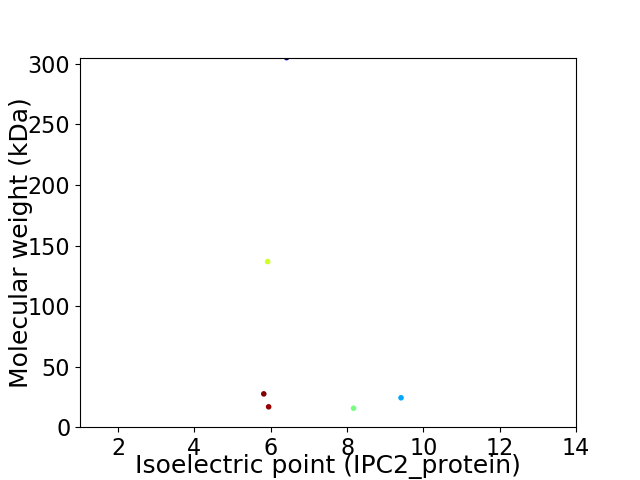
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KJZ1|A0A1L3KJZ1_9VIRU Uncharacterized protein OS=Hubei virga-like virus 23 OX=1923338 PE=4 SV=1
MM1 pKa = 7.27QLILTLTFSLTLIAMTTAQLRR22 pKa = 11.84EE23 pKa = 3.85IFPRR27 pKa = 11.84YY28 pKa = 9.61DD29 pKa = 3.0VARR32 pKa = 11.84GNFEE36 pKa = 5.13DD37 pKa = 5.13DD38 pKa = 3.75ALANLYY44 pKa = 10.61DD45 pKa = 4.74RR46 pKa = 11.84GALTNSADD54 pKa = 3.66INSTCDD60 pKa = 2.87SYY62 pKa = 12.1GCSTEE67 pKa = 4.14FSHH70 pKa = 6.73TFNFNAQRR78 pKa = 11.84ASGTKK83 pKa = 8.86LTLTNNGLQNSTIVISVAEE102 pKa = 3.96ARR104 pKa = 11.84YY105 pKa = 9.57SVSLQYY111 pKa = 11.13LYY113 pKa = 10.42TIYY116 pKa = 9.85PAYY119 pKa = 9.91MFAGWASYY127 pKa = 10.78NYY129 pKa = 10.12EE130 pKa = 4.07EE131 pKa = 5.31NNYY134 pKa = 8.39NQCKK138 pKa = 9.8VKK140 pKa = 10.76YY141 pKa = 9.57DD142 pKa = 3.02WRR144 pKa = 11.84DD145 pKa = 3.15QVNARR150 pKa = 11.84FEE152 pKa = 4.12KK153 pKa = 11.12LEE155 pKa = 4.65DD156 pKa = 3.5YY157 pKa = 10.91VDD159 pKa = 3.1NYY161 pKa = 10.85RR162 pKa = 11.84RR163 pKa = 11.84YY164 pKa = 10.35QIVGSDD170 pKa = 3.5WLRR173 pKa = 11.84VDD175 pKa = 3.38YY176 pKa = 10.74AAWGSVATKK185 pKa = 10.44VCNHH189 pKa = 5.33YY190 pKa = 9.6WATTFTLCQRR200 pKa = 11.84KK201 pKa = 7.06TLVADD206 pKa = 4.69FGEE209 pKa = 5.29GIAVFKK215 pKa = 10.52IAPEE219 pKa = 4.15TTLTMILNIAINDD232 pKa = 3.67DD233 pKa = 3.64VKK235 pKa = 11.23NIEE238 pKa = 4.26VQPSIDD244 pKa = 3.72VPTEE248 pKa = 3.42ITLGDD253 pKa = 3.84LTLSISPAGTIISPLANNYY272 pKa = 9.44YY273 pKa = 10.18LAHH276 pKa = 6.36FQRR279 pKa = 11.84KK280 pKa = 7.39EE281 pKa = 3.8MFDD284 pKa = 3.89TISGAKK290 pKa = 8.19WYY292 pKa = 9.37KK293 pKa = 10.33ANSVPTISSISEE305 pKa = 4.39TITKK309 pKa = 9.23YY310 pKa = 10.41HH311 pKa = 6.42SPNLFPMNAYY321 pKa = 9.65KK322 pKa = 10.14PDD324 pKa = 3.91SEE326 pKa = 4.62TLQQFYY332 pKa = 10.12FDD334 pKa = 4.87KK335 pKa = 10.82MLLQGVNVVKK345 pKa = 10.57CGSSFEE351 pKa = 4.25QEE353 pKa = 3.97ANAKK357 pKa = 10.39NSVQRR362 pKa = 11.84RR363 pKa = 11.84LSKK366 pKa = 10.67LRR368 pKa = 11.84AEE370 pKa = 4.59AAAEE374 pKa = 3.94PSTTKK379 pKa = 9.44MDD381 pKa = 4.5SIISYY386 pKa = 10.61LPTLSNTNPPQTKK399 pKa = 9.2TYY401 pKa = 10.5FNDD404 pKa = 4.23PYY406 pKa = 11.29DD407 pKa = 4.44PYY409 pKa = 11.4DD410 pKa = 3.47NTSISTRR417 pKa = 11.84RR418 pKa = 11.84DD419 pKa = 3.02SDD421 pKa = 3.73CCIPITTEE429 pKa = 3.37SRR431 pKa = 11.84SYY433 pKa = 9.65ITTLLKK439 pKa = 11.03SFDD442 pKa = 4.25LVSDD446 pKa = 3.34IDD448 pKa = 3.74GQYY451 pKa = 11.0EE452 pKa = 3.99VDD454 pKa = 3.69TPSSITQFPGYY465 pKa = 9.99AYY467 pKa = 9.65PNAITMIANNLMKK480 pKa = 10.61VSLSAVLSSQSMRR493 pKa = 11.84IPYY496 pKa = 9.6SAADD500 pKa = 3.72GLVADD505 pKa = 4.11VSIPIMQITTSSAGSYY521 pKa = 10.45FNYY524 pKa = 9.62EE525 pKa = 3.39INYY528 pKa = 9.89VGTANLIPVSARR540 pKa = 11.84HH541 pKa = 5.64INLLKK546 pKa = 9.95RR547 pKa = 11.84TISVTSTGTYY557 pKa = 9.37SGTIAFVSPPDD568 pKa = 3.7PSVDD572 pKa = 3.36LTVCFGSNSVCIKK585 pKa = 10.45PKK587 pKa = 9.02VTNFLTPPQGAVQDD601 pKa = 4.63PDD603 pKa = 3.41TTTCSGVPPSFFSIIITNSGLLAISIIMIIVDD635 pKa = 4.57IILGIVILILLIRR648 pKa = 11.84IVPKK652 pKa = 10.37TFMLFLLLFSTKK664 pKa = 9.46TNALSYY670 pKa = 9.29TAAPISNLTLLDD682 pKa = 4.23NPALDD687 pKa = 3.97NARR690 pKa = 11.84LIYY693 pKa = 10.52SSGISYY699 pKa = 7.65STKK702 pKa = 10.61CEE704 pKa = 3.96TQSCSEE710 pKa = 4.06YY711 pKa = 10.26KK712 pKa = 10.39YY713 pKa = 10.51QSSSPKK719 pKa = 9.21PVEE722 pKa = 3.92YY723 pKa = 10.41TPDD726 pKa = 3.06AKK728 pKa = 10.83MNVYY732 pKa = 10.41LNNYY736 pKa = 8.5AKK738 pKa = 10.82LFAICHH744 pKa = 5.63NFAHH748 pKa = 6.84DD749 pKa = 3.23TGQRR753 pKa = 11.84LPYY756 pKa = 9.28NTVPTPNIDD765 pKa = 3.73LPKK768 pKa = 9.48SWFYY772 pKa = 10.86TIYY775 pKa = 9.84DD776 pKa = 4.0TVDD779 pKa = 3.03SKK781 pKa = 11.56PFVLSLPPYY790 pKa = 10.06LPSTRR795 pKa = 11.84GNYY798 pKa = 7.48VTDD801 pKa = 3.95RR802 pKa = 11.84GLFSPIVSGYY812 pKa = 7.76ATRR815 pKa = 11.84TEE817 pKa = 4.42DD818 pKa = 3.08NTYY821 pKa = 11.63ANITRR826 pKa = 11.84IQPSFQSKK834 pKa = 9.32FLLYY838 pKa = 10.68DD839 pKa = 3.89FYY841 pKa = 11.32NTIYY845 pKa = 10.62GGFMQPYY852 pKa = 8.86SSLIVASQTDD862 pKa = 3.6TTKK865 pKa = 10.7CSNGEE870 pKa = 3.86FTRR873 pKa = 11.84LYY875 pKa = 10.43MMVPSTLISNFTTPIIISDD894 pKa = 3.57IDD896 pKa = 3.97LKK898 pKa = 11.23VAASTYY904 pKa = 10.29RR905 pKa = 11.84AIDD908 pKa = 4.63LISRR912 pKa = 11.84APITYY917 pKa = 10.16GSLALTLRR925 pKa = 11.84NIYY928 pKa = 10.51SGTIEE933 pKa = 4.73ANTFICNTPQSTLYY947 pKa = 10.22RR948 pKa = 11.84ATVVLSCNGVCQTSNTTRR966 pKa = 11.84MLSRR970 pKa = 11.84SVTRR974 pKa = 11.84ADD976 pKa = 3.45RR977 pKa = 11.84TIIDD981 pKa = 5.1DD982 pKa = 3.39IAFYY986 pKa = 7.73TTNAYY991 pKa = 7.49YY992 pKa = 9.89TRR994 pKa = 11.84QGLYY998 pKa = 9.96VPSLVGPIEE1007 pKa = 3.81ITPYY1011 pKa = 10.75SSGSCTVGYY1020 pKa = 10.42DD1021 pKa = 3.67SNGGLLMIDD1030 pKa = 3.75YY1031 pKa = 10.45EE1032 pKa = 4.57DD1033 pKa = 4.04VNVEE1037 pKa = 4.05HH1038 pKa = 6.78VDD1040 pKa = 3.62YY1041 pKa = 11.07RR1042 pKa = 11.84IDD1044 pKa = 3.58LQSFKK1049 pKa = 11.07VSYY1052 pKa = 10.52SPNKK1056 pKa = 9.89VFIKK1060 pKa = 9.91PSPSACTGRR1069 pKa = 11.84TFVSDD1074 pKa = 3.28TNALLTCDD1082 pKa = 4.38SDD1084 pKa = 4.32CCLTTFDD1091 pKa = 4.58NYY1093 pKa = 11.32KK1094 pKa = 10.29QFTIMNAGDD1103 pKa = 3.75VVKK1106 pKa = 10.39FPQSSDD1112 pKa = 2.94ALTLTQYY1119 pKa = 10.32FPSGNQRR1126 pKa = 11.84NVFCPASGCSYY1137 pKa = 10.72ATMLDD1142 pKa = 3.72VYY1144 pKa = 11.23NCALKK1149 pKa = 10.27HH1150 pKa = 5.62HH1151 pKa = 6.65AASTVFLFYY1160 pKa = 9.87FVPIFLGVLIVVYY1173 pKa = 9.95LCRR1176 pKa = 11.84QVIKK1180 pKa = 10.83GFLYY1184 pKa = 10.44GFRR1187 pKa = 11.84NLKK1190 pKa = 9.56FLNPAFYY1197 pKa = 10.66ADD1199 pKa = 4.26KK1200 pKa = 10.38IYY1202 pKa = 10.81NPSVAAVSQIRR1213 pKa = 11.84NSTLRR1218 pKa = 11.84RR1219 pKa = 11.84VKK1221 pKa = 10.48RR1222 pKa = 11.84ITSNKK1227 pKa = 9.64DD1228 pKa = 2.62AA1229 pKa = 4.99
MM1 pKa = 7.27QLILTLTFSLTLIAMTTAQLRR22 pKa = 11.84EE23 pKa = 3.85IFPRR27 pKa = 11.84YY28 pKa = 9.61DD29 pKa = 3.0VARR32 pKa = 11.84GNFEE36 pKa = 5.13DD37 pKa = 5.13DD38 pKa = 3.75ALANLYY44 pKa = 10.61DD45 pKa = 4.74RR46 pKa = 11.84GALTNSADD54 pKa = 3.66INSTCDD60 pKa = 2.87SYY62 pKa = 12.1GCSTEE67 pKa = 4.14FSHH70 pKa = 6.73TFNFNAQRR78 pKa = 11.84ASGTKK83 pKa = 8.86LTLTNNGLQNSTIVISVAEE102 pKa = 3.96ARR104 pKa = 11.84YY105 pKa = 9.57SVSLQYY111 pKa = 11.13LYY113 pKa = 10.42TIYY116 pKa = 9.85PAYY119 pKa = 9.91MFAGWASYY127 pKa = 10.78NYY129 pKa = 10.12EE130 pKa = 4.07EE131 pKa = 5.31NNYY134 pKa = 8.39NQCKK138 pKa = 9.8VKK140 pKa = 10.76YY141 pKa = 9.57DD142 pKa = 3.02WRR144 pKa = 11.84DD145 pKa = 3.15QVNARR150 pKa = 11.84FEE152 pKa = 4.12KK153 pKa = 11.12LEE155 pKa = 4.65DD156 pKa = 3.5YY157 pKa = 10.91VDD159 pKa = 3.1NYY161 pKa = 10.85RR162 pKa = 11.84RR163 pKa = 11.84YY164 pKa = 10.35QIVGSDD170 pKa = 3.5WLRR173 pKa = 11.84VDD175 pKa = 3.38YY176 pKa = 10.74AAWGSVATKK185 pKa = 10.44VCNHH189 pKa = 5.33YY190 pKa = 9.6WATTFTLCQRR200 pKa = 11.84KK201 pKa = 7.06TLVADD206 pKa = 4.69FGEE209 pKa = 5.29GIAVFKK215 pKa = 10.52IAPEE219 pKa = 4.15TTLTMILNIAINDD232 pKa = 3.67DD233 pKa = 3.64VKK235 pKa = 11.23NIEE238 pKa = 4.26VQPSIDD244 pKa = 3.72VPTEE248 pKa = 3.42ITLGDD253 pKa = 3.84LTLSISPAGTIISPLANNYY272 pKa = 9.44YY273 pKa = 10.18LAHH276 pKa = 6.36FQRR279 pKa = 11.84KK280 pKa = 7.39EE281 pKa = 3.8MFDD284 pKa = 3.89TISGAKK290 pKa = 8.19WYY292 pKa = 9.37KK293 pKa = 10.33ANSVPTISSISEE305 pKa = 4.39TITKK309 pKa = 9.23YY310 pKa = 10.41HH311 pKa = 6.42SPNLFPMNAYY321 pKa = 9.65KK322 pKa = 10.14PDD324 pKa = 3.91SEE326 pKa = 4.62TLQQFYY332 pKa = 10.12FDD334 pKa = 4.87KK335 pKa = 10.82MLLQGVNVVKK345 pKa = 10.57CGSSFEE351 pKa = 4.25QEE353 pKa = 3.97ANAKK357 pKa = 10.39NSVQRR362 pKa = 11.84RR363 pKa = 11.84LSKK366 pKa = 10.67LRR368 pKa = 11.84AEE370 pKa = 4.59AAAEE374 pKa = 3.94PSTTKK379 pKa = 9.44MDD381 pKa = 4.5SIISYY386 pKa = 10.61LPTLSNTNPPQTKK399 pKa = 9.2TYY401 pKa = 10.5FNDD404 pKa = 4.23PYY406 pKa = 11.29DD407 pKa = 4.44PYY409 pKa = 11.4DD410 pKa = 3.47NTSISTRR417 pKa = 11.84RR418 pKa = 11.84DD419 pKa = 3.02SDD421 pKa = 3.73CCIPITTEE429 pKa = 3.37SRR431 pKa = 11.84SYY433 pKa = 9.65ITTLLKK439 pKa = 11.03SFDD442 pKa = 4.25LVSDD446 pKa = 3.34IDD448 pKa = 3.74GQYY451 pKa = 11.0EE452 pKa = 3.99VDD454 pKa = 3.69TPSSITQFPGYY465 pKa = 9.99AYY467 pKa = 9.65PNAITMIANNLMKK480 pKa = 10.61VSLSAVLSSQSMRR493 pKa = 11.84IPYY496 pKa = 9.6SAADD500 pKa = 3.72GLVADD505 pKa = 4.11VSIPIMQITTSSAGSYY521 pKa = 10.45FNYY524 pKa = 9.62EE525 pKa = 3.39INYY528 pKa = 9.89VGTANLIPVSARR540 pKa = 11.84HH541 pKa = 5.64INLLKK546 pKa = 9.95RR547 pKa = 11.84TISVTSTGTYY557 pKa = 9.37SGTIAFVSPPDD568 pKa = 3.7PSVDD572 pKa = 3.36LTVCFGSNSVCIKK585 pKa = 10.45PKK587 pKa = 9.02VTNFLTPPQGAVQDD601 pKa = 4.63PDD603 pKa = 3.41TTTCSGVPPSFFSIIITNSGLLAISIIMIIVDD635 pKa = 4.57IILGIVILILLIRR648 pKa = 11.84IVPKK652 pKa = 10.37TFMLFLLLFSTKK664 pKa = 9.46TNALSYY670 pKa = 9.29TAAPISNLTLLDD682 pKa = 4.23NPALDD687 pKa = 3.97NARR690 pKa = 11.84LIYY693 pKa = 10.52SSGISYY699 pKa = 7.65STKK702 pKa = 10.61CEE704 pKa = 3.96TQSCSEE710 pKa = 4.06YY711 pKa = 10.26KK712 pKa = 10.39YY713 pKa = 10.51QSSSPKK719 pKa = 9.21PVEE722 pKa = 3.92YY723 pKa = 10.41TPDD726 pKa = 3.06AKK728 pKa = 10.83MNVYY732 pKa = 10.41LNNYY736 pKa = 8.5AKK738 pKa = 10.82LFAICHH744 pKa = 5.63NFAHH748 pKa = 6.84DD749 pKa = 3.23TGQRR753 pKa = 11.84LPYY756 pKa = 9.28NTVPTPNIDD765 pKa = 3.73LPKK768 pKa = 9.48SWFYY772 pKa = 10.86TIYY775 pKa = 9.84DD776 pKa = 4.0TVDD779 pKa = 3.03SKK781 pKa = 11.56PFVLSLPPYY790 pKa = 10.06LPSTRR795 pKa = 11.84GNYY798 pKa = 7.48VTDD801 pKa = 3.95RR802 pKa = 11.84GLFSPIVSGYY812 pKa = 7.76ATRR815 pKa = 11.84TEE817 pKa = 4.42DD818 pKa = 3.08NTYY821 pKa = 11.63ANITRR826 pKa = 11.84IQPSFQSKK834 pKa = 9.32FLLYY838 pKa = 10.68DD839 pKa = 3.89FYY841 pKa = 11.32NTIYY845 pKa = 10.62GGFMQPYY852 pKa = 8.86SSLIVASQTDD862 pKa = 3.6TTKK865 pKa = 10.7CSNGEE870 pKa = 3.86FTRR873 pKa = 11.84LYY875 pKa = 10.43MMVPSTLISNFTTPIIISDD894 pKa = 3.57IDD896 pKa = 3.97LKK898 pKa = 11.23VAASTYY904 pKa = 10.29RR905 pKa = 11.84AIDD908 pKa = 4.63LISRR912 pKa = 11.84APITYY917 pKa = 10.16GSLALTLRR925 pKa = 11.84NIYY928 pKa = 10.51SGTIEE933 pKa = 4.73ANTFICNTPQSTLYY947 pKa = 10.22RR948 pKa = 11.84ATVVLSCNGVCQTSNTTRR966 pKa = 11.84MLSRR970 pKa = 11.84SVTRR974 pKa = 11.84ADD976 pKa = 3.45RR977 pKa = 11.84TIIDD981 pKa = 5.1DD982 pKa = 3.39IAFYY986 pKa = 7.73TTNAYY991 pKa = 7.49YY992 pKa = 9.89TRR994 pKa = 11.84QGLYY998 pKa = 9.96VPSLVGPIEE1007 pKa = 3.81ITPYY1011 pKa = 10.75SSGSCTVGYY1020 pKa = 10.42DD1021 pKa = 3.67SNGGLLMIDD1030 pKa = 3.75YY1031 pKa = 10.45EE1032 pKa = 4.57DD1033 pKa = 4.04VNVEE1037 pKa = 4.05HH1038 pKa = 6.78VDD1040 pKa = 3.62YY1041 pKa = 11.07RR1042 pKa = 11.84IDD1044 pKa = 3.58LQSFKK1049 pKa = 11.07VSYY1052 pKa = 10.52SPNKK1056 pKa = 9.89VFIKK1060 pKa = 9.91PSPSACTGRR1069 pKa = 11.84TFVSDD1074 pKa = 3.28TNALLTCDD1082 pKa = 4.38SDD1084 pKa = 4.32CCLTTFDD1091 pKa = 4.58NYY1093 pKa = 11.32KK1094 pKa = 10.29QFTIMNAGDD1103 pKa = 3.75VVKK1106 pKa = 10.39FPQSSDD1112 pKa = 2.94ALTLTQYY1119 pKa = 10.32FPSGNQRR1126 pKa = 11.84NVFCPASGCSYY1137 pKa = 10.72ATMLDD1142 pKa = 3.72VYY1144 pKa = 11.23NCALKK1149 pKa = 10.27HH1150 pKa = 5.62HH1151 pKa = 6.65AASTVFLFYY1160 pKa = 9.87FVPIFLGVLIVVYY1173 pKa = 9.95LCRR1176 pKa = 11.84QVIKK1180 pKa = 10.83GFLYY1184 pKa = 10.44GFRR1187 pKa = 11.84NLKK1190 pKa = 9.56FLNPAFYY1197 pKa = 10.66ADD1199 pKa = 4.26KK1200 pKa = 10.38IYY1202 pKa = 10.81NPSVAAVSQIRR1213 pKa = 11.84NSTLRR1218 pKa = 11.84RR1219 pKa = 11.84VKK1221 pKa = 10.48RR1222 pKa = 11.84ITSNKK1227 pKa = 9.64DD1228 pKa = 2.62AA1229 pKa = 4.99
Molecular weight: 136.75 kDa
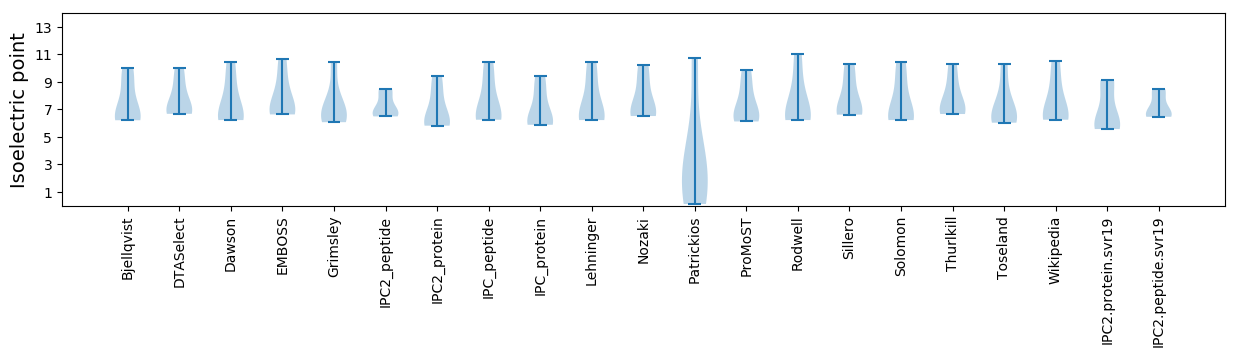
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJS4|A0A1L3KJS4_9VIRU Putative glycoprotein OS=Hubei virga-like virus 23 OX=1923338 PE=4 SV=1
MM1 pKa = 7.03NRR3 pKa = 11.84SVSTQQLDD11 pKa = 3.47RR12 pKa = 11.84FRR14 pKa = 11.84PNARR18 pKa = 11.84RR19 pKa = 11.84GQTRR23 pKa = 11.84RR24 pKa = 11.84SSSLSRR30 pKa = 11.84NDD32 pKa = 2.79NPAYY36 pKa = 9.87YY37 pKa = 9.96KK38 pKa = 10.69PKK40 pKa = 9.71TPPRR44 pKa = 11.84NNVKK48 pKa = 9.7VVYY51 pKa = 10.33LSEE54 pKa = 3.8PTANIKK60 pKa = 9.18TKK62 pKa = 10.48FNGYY66 pKa = 9.27VNYY69 pKa = 9.76PDD71 pKa = 6.43LFIKK75 pKa = 9.49WQDD78 pKa = 3.28LVSIVLRR85 pKa = 11.84TAKK88 pKa = 10.11IFSSKK93 pKa = 8.96LTNKK97 pKa = 10.11DD98 pKa = 3.15FARR101 pKa = 11.84LFTLADD107 pKa = 3.94RR108 pKa = 11.84SIKK111 pKa = 10.67LHH113 pKa = 4.66TTCNYY118 pKa = 9.35VFEE121 pKa = 4.72IQGQSDD127 pKa = 3.2KK128 pKa = 10.75GAKK131 pKa = 9.7VMTIAMHH138 pKa = 6.65GPASANDD145 pKa = 3.67LFINDD150 pKa = 4.3SLNDD154 pKa = 3.72IIAAVEE160 pKa = 4.28LIGAQTIIPQTLSSGKK176 pKa = 10.2FFFTKK181 pKa = 10.48VKK183 pKa = 10.55LNSNSKK189 pKa = 10.11EE190 pKa = 3.9SSIVNDD196 pKa = 3.81KK197 pKa = 10.99NSPSLFSDD205 pKa = 4.23TIKK208 pKa = 10.88LGLSSGSSKK217 pKa = 11.03SS218 pKa = 3.32
MM1 pKa = 7.03NRR3 pKa = 11.84SVSTQQLDD11 pKa = 3.47RR12 pKa = 11.84FRR14 pKa = 11.84PNARR18 pKa = 11.84RR19 pKa = 11.84GQTRR23 pKa = 11.84RR24 pKa = 11.84SSSLSRR30 pKa = 11.84NDD32 pKa = 2.79NPAYY36 pKa = 9.87YY37 pKa = 9.96KK38 pKa = 10.69PKK40 pKa = 9.71TPPRR44 pKa = 11.84NNVKK48 pKa = 9.7VVYY51 pKa = 10.33LSEE54 pKa = 3.8PTANIKK60 pKa = 9.18TKK62 pKa = 10.48FNGYY66 pKa = 9.27VNYY69 pKa = 9.76PDD71 pKa = 6.43LFIKK75 pKa = 9.49WQDD78 pKa = 3.28LVSIVLRR85 pKa = 11.84TAKK88 pKa = 10.11IFSSKK93 pKa = 8.96LTNKK97 pKa = 10.11DD98 pKa = 3.15FARR101 pKa = 11.84LFTLADD107 pKa = 3.94RR108 pKa = 11.84SIKK111 pKa = 10.67LHH113 pKa = 4.66TTCNYY118 pKa = 9.35VFEE121 pKa = 4.72IQGQSDD127 pKa = 3.2KK128 pKa = 10.75GAKK131 pKa = 9.7VMTIAMHH138 pKa = 6.65GPASANDD145 pKa = 3.67LFINDD150 pKa = 4.3SLNDD154 pKa = 3.72IIAAVEE160 pKa = 4.28LIGAQTIIPQTLSSGKK176 pKa = 10.2FFFTKK181 pKa = 10.48VKK183 pKa = 10.55LNSNSKK189 pKa = 10.11EE190 pKa = 3.9SSIVNDD196 pKa = 3.81KK197 pKa = 10.99NSPSLFSDD205 pKa = 4.23TIKK208 pKa = 10.88LGLSSGSSKK217 pKa = 11.03SS218 pKa = 3.32
Molecular weight: 24.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4680 |
136 |
2702 |
780.0 |
87.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.641 ± 0.463 | 2.521 ± 0.21 |
5.726 ± 0.477 | 4.081 ± 0.993 |
4.487 ± 0.342 | 4.252 ± 0.288 |
2.244 ± 0.564 | 7.692 ± 0.543 |
5.62 ± 0.949 | 8.611 ± 0.441 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.902 ± 0.268 | 5.897 ± 0.473 |
4.444 ± 0.521 | 3.12 ± 0.254 |
4.252 ± 0.213 | 8.974 ± 0.874 |
7.436 ± 0.944 | 7.415 ± 0.717 |
0.577 ± 0.164 | 5.064 ± 0.635 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
