
Mesorhizobium sp. Root157
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Mesorhizobium; unclassified Mesorhizobium
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
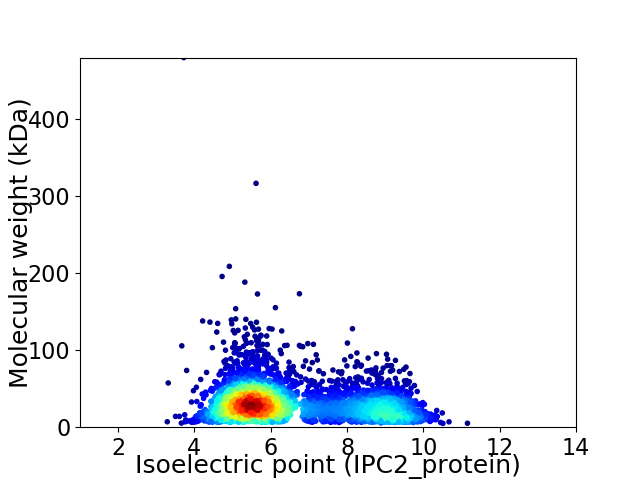
Virtual 2D-PAGE plot for 3925 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q8AFI2|A0A0Q8AFI2_9RHIZ ATPase OS=Mesorhizobium sp. Root157 OX=1736477 GN=ASD64_10455 PE=4 SV=1
MM1 pKa = 7.98DD2 pKa = 5.04KK3 pKa = 9.88VTDD6 pKa = 4.55SIAHH10 pKa = 6.64FIGLFQIAAEE20 pKa = 4.12QARR23 pKa = 11.84MRR25 pKa = 11.84DD26 pKa = 3.54DD27 pKa = 3.37YY28 pKa = 11.93LEE30 pKa = 4.32FVARR34 pKa = 11.84RR35 pKa = 11.84AHH37 pKa = 7.28DD38 pKa = 3.64EE39 pKa = 4.39APPSDD44 pKa = 4.04EE45 pKa = 3.81PVQIKK50 pKa = 9.98FDD52 pKa = 3.57APYY55 pKa = 11.19ALDD58 pKa = 3.7NPEE61 pKa = 4.41PGIHH65 pKa = 5.88YY66 pKa = 10.44VPAPNSIEE74 pKa = 4.23PYY76 pKa = 8.55VAHH79 pKa = 7.13HH80 pKa = 6.72EE81 pKa = 4.11PLYY84 pKa = 10.38AAPEE88 pKa = 4.15IPISAQMQPVIYY100 pKa = 8.94PGYY103 pKa = 8.08VPPVFLHH110 pKa = 6.3HH111 pKa = 6.94ASSSLKK117 pKa = 10.4IIDD120 pKa = 4.05PQPPGSVASIVSQTNHH136 pKa = 6.64LSDD139 pKa = 3.61NDD141 pKa = 3.65FVNVGGSDD149 pKa = 3.25AVFEE153 pKa = 4.72QIGTPQTSLQEE164 pKa = 4.42LVDD167 pKa = 3.88QASQAMPILAGSLPGSSQDD186 pKa = 2.46IGTFITTSADD196 pKa = 3.65SIHH199 pKa = 7.3DD200 pKa = 3.72FLAGLEE206 pKa = 4.25SGQQAGEE213 pKa = 4.23AGSSQTIVDD222 pKa = 4.04NGDD225 pKa = 3.4GTSTATQVTAVTPAEE240 pKa = 3.9DD241 pKa = 3.21TVYY244 pKa = 11.32VNGTATDD251 pKa = 3.86EE252 pKa = 4.13APKK255 pKa = 10.64LDD257 pKa = 4.99DD258 pKa = 4.21YY259 pKa = 11.8LPAASPLVEE268 pKa = 4.4DD269 pKa = 5.06TPPEE273 pKa = 4.21AAPEE277 pKa = 4.02ASNGSWQTGSTATGQVVHH295 pKa = 6.35GQGSIAVTASVEE307 pKa = 3.79ISTGLNVLVNSATLVSDD324 pKa = 3.38TLEE327 pKa = 3.94GHH329 pKa = 6.15VFAIAGDD336 pKa = 4.33HH337 pKa = 6.58ISLNLIVQINAWSDD351 pKa = 3.22ADD353 pKa = 3.88SVGASMSGWNCPPDD367 pKa = 3.36GTTAFNIASMEE378 pKa = 4.04HH379 pKa = 6.18VEE381 pKa = 4.76LNTGTDD387 pKa = 3.63EE388 pKa = 4.4PSGAGTPVFPKK399 pKa = 9.8AWAVTEE405 pKa = 3.92ITGDD409 pKa = 4.33FISLNWLQQLNFVIDD424 pKa = 3.65NDD426 pKa = 4.32TVVASSSAGVTSMVGTGANQTFNSLSIFDD455 pKa = 3.93LGKK458 pKa = 10.71YY459 pKa = 10.0FDD461 pKa = 5.93LILIGGNYY469 pKa = 9.29YY470 pKa = 10.07NANIILQKK478 pKa = 11.08NIMLDD483 pKa = 3.51DD484 pKa = 5.51DD485 pKa = 3.94IVGGVSGFQTSGPGAVSTSDD505 pKa = 3.15NLLWNEE511 pKa = 4.1ARR513 pKa = 11.84ITNIGTANTDD523 pKa = 3.2GMPDD527 pKa = 3.48GFHH530 pKa = 6.64NALNEE535 pKa = 3.8FAAGNKK541 pKa = 9.01VLSADD546 pKa = 3.88VLNDD550 pKa = 3.29SAFAGLAGLHH560 pKa = 5.61VLYY563 pKa = 10.29ISGSIYY569 pKa = 10.41DD570 pKa = 3.92LQYY573 pKa = 10.64IQQTNILGDD582 pKa = 3.91ADD584 pKa = 3.66QVAAAMQGVNPTSDD598 pKa = 4.15ANWSITTGSNVLVNQAQIIDD618 pKa = 4.08VGPGGAIYY626 pKa = 10.56YY627 pKa = 10.7GGGQYY632 pKa = 10.51SDD634 pKa = 4.14EE635 pKa = 4.99LLVQTDD641 pKa = 4.53IIRR644 pKa = 11.84TDD646 pKa = 2.81NGTDD650 pKa = 3.21VKK652 pKa = 11.44SPDD655 pKa = 3.4QLVNEE660 pKa = 4.55AVAFLSDD667 pKa = 5.55DD668 pKa = 3.75MLTPDD673 pKa = 4.44QSQEE677 pKa = 3.73HH678 pKa = 4.98QMGIKK683 pKa = 10.06DD684 pKa = 4.1DD685 pKa = 4.13VGMHH689 pKa = 6.8PAHH692 pKa = 6.59TDD694 pKa = 2.51IMQSVVTT701 pKa = 4.53
MM1 pKa = 7.98DD2 pKa = 5.04KK3 pKa = 9.88VTDD6 pKa = 4.55SIAHH10 pKa = 6.64FIGLFQIAAEE20 pKa = 4.12QARR23 pKa = 11.84MRR25 pKa = 11.84DD26 pKa = 3.54DD27 pKa = 3.37YY28 pKa = 11.93LEE30 pKa = 4.32FVARR34 pKa = 11.84RR35 pKa = 11.84AHH37 pKa = 7.28DD38 pKa = 3.64EE39 pKa = 4.39APPSDD44 pKa = 4.04EE45 pKa = 3.81PVQIKK50 pKa = 9.98FDD52 pKa = 3.57APYY55 pKa = 11.19ALDD58 pKa = 3.7NPEE61 pKa = 4.41PGIHH65 pKa = 5.88YY66 pKa = 10.44VPAPNSIEE74 pKa = 4.23PYY76 pKa = 8.55VAHH79 pKa = 7.13HH80 pKa = 6.72EE81 pKa = 4.11PLYY84 pKa = 10.38AAPEE88 pKa = 4.15IPISAQMQPVIYY100 pKa = 8.94PGYY103 pKa = 8.08VPPVFLHH110 pKa = 6.3HH111 pKa = 6.94ASSSLKK117 pKa = 10.4IIDD120 pKa = 4.05PQPPGSVASIVSQTNHH136 pKa = 6.64LSDD139 pKa = 3.61NDD141 pKa = 3.65FVNVGGSDD149 pKa = 3.25AVFEE153 pKa = 4.72QIGTPQTSLQEE164 pKa = 4.42LVDD167 pKa = 3.88QASQAMPILAGSLPGSSQDD186 pKa = 2.46IGTFITTSADD196 pKa = 3.65SIHH199 pKa = 7.3DD200 pKa = 3.72FLAGLEE206 pKa = 4.25SGQQAGEE213 pKa = 4.23AGSSQTIVDD222 pKa = 4.04NGDD225 pKa = 3.4GTSTATQVTAVTPAEE240 pKa = 3.9DD241 pKa = 3.21TVYY244 pKa = 11.32VNGTATDD251 pKa = 3.86EE252 pKa = 4.13APKK255 pKa = 10.64LDD257 pKa = 4.99DD258 pKa = 4.21YY259 pKa = 11.8LPAASPLVEE268 pKa = 4.4DD269 pKa = 5.06TPPEE273 pKa = 4.21AAPEE277 pKa = 4.02ASNGSWQTGSTATGQVVHH295 pKa = 6.35GQGSIAVTASVEE307 pKa = 3.79ISTGLNVLVNSATLVSDD324 pKa = 3.38TLEE327 pKa = 3.94GHH329 pKa = 6.15VFAIAGDD336 pKa = 4.33HH337 pKa = 6.58ISLNLIVQINAWSDD351 pKa = 3.22ADD353 pKa = 3.88SVGASMSGWNCPPDD367 pKa = 3.36GTTAFNIASMEE378 pKa = 4.04HH379 pKa = 6.18VEE381 pKa = 4.76LNTGTDD387 pKa = 3.63EE388 pKa = 4.4PSGAGTPVFPKK399 pKa = 9.8AWAVTEE405 pKa = 3.92ITGDD409 pKa = 4.33FISLNWLQQLNFVIDD424 pKa = 3.65NDD426 pKa = 4.32TVVASSSAGVTSMVGTGANQTFNSLSIFDD455 pKa = 3.93LGKK458 pKa = 10.71YY459 pKa = 10.0FDD461 pKa = 5.93LILIGGNYY469 pKa = 9.29YY470 pKa = 10.07NANIILQKK478 pKa = 11.08NIMLDD483 pKa = 3.51DD484 pKa = 5.51DD485 pKa = 3.94IVGGVSGFQTSGPGAVSTSDD505 pKa = 3.15NLLWNEE511 pKa = 4.1ARR513 pKa = 11.84ITNIGTANTDD523 pKa = 3.2GMPDD527 pKa = 3.48GFHH530 pKa = 6.64NALNEE535 pKa = 3.8FAAGNKK541 pKa = 9.01VLSADD546 pKa = 3.88VLNDD550 pKa = 3.29SAFAGLAGLHH560 pKa = 5.61VLYY563 pKa = 10.29ISGSIYY569 pKa = 10.41DD570 pKa = 3.92LQYY573 pKa = 10.64IQQTNILGDD582 pKa = 3.91ADD584 pKa = 3.66QVAAAMQGVNPTSDD598 pKa = 4.15ANWSITTGSNVLVNQAQIIDD618 pKa = 4.08VGPGGAIYY626 pKa = 10.56YY627 pKa = 10.7GGGQYY632 pKa = 10.51SDD634 pKa = 4.14EE635 pKa = 4.99LLVQTDD641 pKa = 4.53IIRR644 pKa = 11.84TDD646 pKa = 2.81NGTDD650 pKa = 3.21VKK652 pKa = 11.44SPDD655 pKa = 3.4QLVNEE660 pKa = 4.55AVAFLSDD667 pKa = 5.55DD668 pKa = 3.75MLTPDD673 pKa = 4.44QSQEE677 pKa = 3.73HH678 pKa = 4.98QMGIKK683 pKa = 10.06DD684 pKa = 4.1DD685 pKa = 4.13VGMHH689 pKa = 6.8PAHH692 pKa = 6.59TDD694 pKa = 2.51IMQSVVTT701 pKa = 4.53
Molecular weight: 73.61 kDa
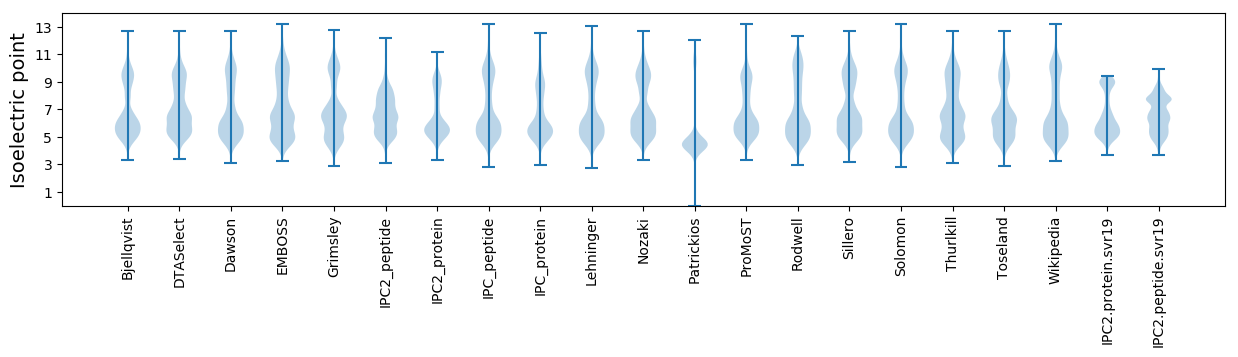
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q8AWM5|A0A0Q8AWM5_9RHIZ Myo-inosose-2 dehydratase OS=Mesorhizobium sp. Root157 OX=1736477 GN=ASD64_02730 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.14GGRR28 pKa = 11.84SVVAARR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84QRR41 pKa = 11.84LSAA44 pKa = 4.11
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.14GGRR28 pKa = 11.84SVVAARR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84QRR41 pKa = 11.84LSAA44 pKa = 4.11
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1190505 |
41 |
4699 |
303.3 |
32.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.313 ± 0.056 | 0.788 ± 0.012 |
5.755 ± 0.035 | 5.792 ± 0.04 |
3.86 ± 0.031 | 8.552 ± 0.047 |
2.011 ± 0.018 | 5.427 ± 0.029 |
3.872 ± 0.033 | 9.725 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.565 ± 0.019 | 2.762 ± 0.026 |
4.875 ± 0.027 | 3.092 ± 0.023 |
6.758 ± 0.045 | 5.555 ± 0.03 |
5.253 ± 0.035 | 7.5 ± 0.029 |
1.287 ± 0.017 | 2.256 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
