
Infectious hypodermal and hematopoietic necrosis virus (IHHNV) (Decapod penstyldensovirus 1)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Quintoviricetes; Piccovirales; Parvoviridae; Densovirinae; Penstyldensovirus
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
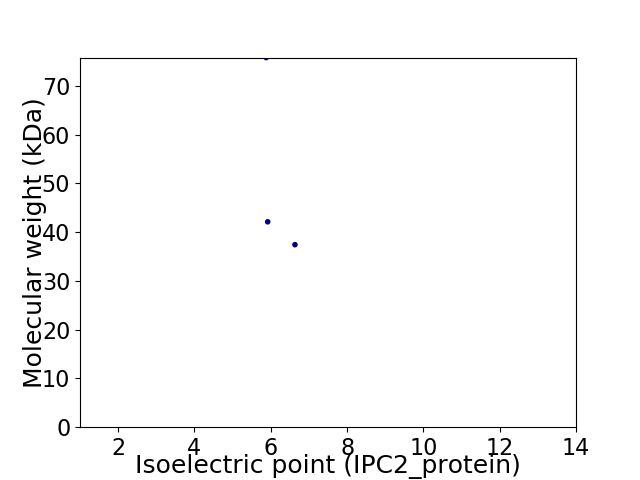
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9IBP6|Q9IBP6_IHHNV Non-structural protein OS=Infectious hypodermal and hematopoietic necrosis virus OX=1513224 PE=4 SV=1
MM1 pKa = 7.61AKK3 pKa = 10.33DD4 pKa = 3.34ILHH7 pKa = 5.97TRR9 pKa = 11.84QGEE12 pKa = 4.22PEE14 pKa = 4.3SLSEE18 pKa = 4.48LLRR21 pKa = 11.84EE22 pKa = 4.39RR23 pKa = 11.84TNEE26 pKa = 3.91THH28 pKa = 6.69SSQGISPSLLTPGPNQDD45 pKa = 3.87PKK47 pKa = 10.47PTTEE51 pKa = 3.69QLLNMSEE58 pKa = 4.49EE59 pKa = 4.32LFQFSDD65 pKa = 4.19EE66 pKa = 4.49EE67 pKa = 4.99DD68 pKa = 3.55NSQTPPRR75 pKa = 11.84TSTPEE80 pKa = 3.48QTDD83 pKa = 3.3PKK85 pKa = 10.77VCVDD89 pKa = 3.31NLGIRR94 pKa = 11.84EE95 pKa = 4.31GTGNGTIQLGSEE107 pKa = 4.5SEE109 pKa = 4.25TSLGSVGNSNDD120 pKa = 3.5RR121 pKa = 11.84GKK123 pKa = 10.64KK124 pKa = 6.8RR125 pKa = 11.84QRR127 pKa = 11.84GITYY131 pKa = 10.1ISDD134 pKa = 3.57TSDD137 pKa = 2.95SSGSDD142 pKa = 3.58DD143 pKa = 4.09EE144 pKa = 5.38NLGTPHH150 pKa = 6.87RR151 pKa = 11.84NKK153 pKa = 9.66RR154 pKa = 11.84TRR156 pKa = 11.84NSNTTKK162 pKa = 9.75EE163 pKa = 4.33TGGGDD168 pKa = 3.37NRR170 pKa = 11.84RR171 pKa = 11.84HH172 pKa = 5.48QEE174 pKa = 3.68SDD176 pKa = 2.77HH177 pKa = 6.22GSNGNRR183 pKa = 11.84LEE185 pKa = 4.2PTDD188 pKa = 4.16GGEE191 pKa = 4.31SCSSGTQPDD200 pKa = 5.02FIEE203 pKa = 4.72GTPNGPDD210 pKa = 3.15EE211 pKa = 4.25MDD213 pKa = 3.0GRR215 pKa = 11.84RR216 pKa = 11.84LEE218 pKa = 3.88EE219 pKa = 4.15SEE221 pKa = 3.86IDD223 pKa = 3.57KK224 pKa = 10.85QVEE227 pKa = 4.15STTWYY232 pKa = 9.04TFVIRR237 pKa = 11.84EE238 pKa = 4.04KK239 pKa = 9.67PQPRR243 pKa = 11.84RR244 pKa = 11.84LSGRR248 pKa = 11.84TPNFTITDD256 pKa = 4.0HH257 pKa = 6.73GDD259 pKa = 2.86HH260 pKa = 5.68WHH262 pKa = 6.06ITYY265 pKa = 10.5SGHH268 pKa = 5.99PTNKK272 pKa = 7.59TRR274 pKa = 11.84HH275 pKa = 5.82RR276 pKa = 11.84ATILAYY282 pKa = 10.75LGVTFAARR290 pKa = 11.84AEE292 pKa = 4.22AEE294 pKa = 4.06ATTVLVRR301 pKa = 11.84NIKK304 pKa = 10.03RR305 pKa = 11.84WILYY309 pKa = 8.45LIRR312 pKa = 11.84YY313 pKa = 7.88GIEE316 pKa = 3.49RR317 pKa = 11.84LSYY320 pKa = 10.4FGLGHH325 pKa = 7.53AIFKK329 pKa = 10.55RR330 pKa = 11.84IIKK333 pKa = 9.25YY334 pKa = 7.27FQQYY338 pKa = 9.86RR339 pKa = 11.84RR340 pKa = 11.84DD341 pKa = 3.56EE342 pKa = 4.39DD343 pKa = 4.77AVDD346 pKa = 4.96GPCPYY351 pKa = 8.15MTTTRR356 pKa = 11.84EE357 pKa = 3.77DD358 pKa = 3.41RR359 pKa = 11.84AEE361 pKa = 4.17EE362 pKa = 4.12KK363 pKa = 10.47PKK365 pKa = 10.81EE366 pKa = 3.92NSAEE370 pKa = 3.88YY371 pKa = 10.14DD372 pKa = 3.78YY373 pKa = 11.29LQHH376 pKa = 6.98LVKK379 pKa = 9.86TKK381 pKa = 9.44SARR384 pKa = 11.84TVQEE388 pKa = 4.28LVNKK392 pKa = 10.41LDD394 pKa = 3.88DD395 pKa = 4.09EE396 pKa = 4.77EE397 pKa = 5.64YY398 pKa = 9.93KK399 pKa = 10.68QLWTRR404 pKa = 11.84TRR406 pKa = 11.84GQYY409 pKa = 9.99KK410 pKa = 10.45DD411 pKa = 3.35KK412 pKa = 11.13LRR414 pKa = 11.84GILTYY419 pKa = 11.05YY420 pKa = 10.42NNKK423 pKa = 9.52KK424 pKa = 10.26KK425 pKa = 10.87SNQSQLSLITNLQNISKK442 pKa = 10.41RR443 pKa = 11.84KK444 pKa = 8.29PDD446 pKa = 3.96YY447 pKa = 11.73DD448 pKa = 3.19NMQWIKK454 pKa = 11.46YY455 pKa = 7.86MLANNDD461 pKa = 2.63IRR463 pKa = 11.84VPEE466 pKa = 3.87ILAWIIIVADD476 pKa = 3.85KK477 pKa = 11.04KK478 pKa = 10.74LDD480 pKa = 4.15KK481 pKa = 11.06INTLVLQGPTGTGKK495 pKa = 10.59SLTIGALLGKK505 pKa = 10.23LNTGLVTRR513 pKa = 11.84TGDD516 pKa = 3.32SNTFHH521 pKa = 6.75LQNLIGKK528 pKa = 9.11SYY530 pKa = 11.56ALFEE534 pKa = 4.41EE535 pKa = 4.68PRR537 pKa = 11.84ISQITVDD544 pKa = 4.01DD545 pKa = 4.36FKK547 pKa = 11.82LLFEE551 pKa = 4.99GSDD554 pKa = 3.38LEE556 pKa = 4.9VNIKK560 pKa = 9.77HH561 pKa = 5.57QEE563 pKa = 3.88SEE565 pKa = 3.78IMGRR569 pKa = 11.84IPIFISTNKK578 pKa = 10.59DD579 pKa = 2.13IDD581 pKa = 3.87YY582 pKa = 8.44WVPPADD588 pKa = 3.56GKK590 pKa = 10.96ALQTRR595 pKa = 11.84TKK597 pKa = 9.22TFHH600 pKa = 6.65LTRR603 pKa = 11.84QIKK606 pKa = 10.09GLSDD610 pKa = 3.52RR611 pKa = 11.84MNSQYY616 pKa = 11.04DD617 pKa = 3.63INPPPDD623 pKa = 5.19KK624 pKa = 10.18ITSDD628 pKa = 3.66DD629 pKa = 3.76FLGLFQEE636 pKa = 4.62YY637 pKa = 9.77EE638 pKa = 4.23KK639 pKa = 10.94EE640 pKa = 3.65IDD642 pKa = 4.68DD643 pKa = 5.48IIDD646 pKa = 3.14NHH648 pKa = 5.65VRR650 pKa = 11.84RR651 pKa = 11.84FNKK654 pKa = 10.1SKK656 pKa = 10.22PKK658 pKa = 10.48EE659 pKa = 4.2KK660 pKa = 10.03IQEE663 pKa = 4.24GCTT666 pKa = 3.34
MM1 pKa = 7.61AKK3 pKa = 10.33DD4 pKa = 3.34ILHH7 pKa = 5.97TRR9 pKa = 11.84QGEE12 pKa = 4.22PEE14 pKa = 4.3SLSEE18 pKa = 4.48LLRR21 pKa = 11.84EE22 pKa = 4.39RR23 pKa = 11.84TNEE26 pKa = 3.91THH28 pKa = 6.69SSQGISPSLLTPGPNQDD45 pKa = 3.87PKK47 pKa = 10.47PTTEE51 pKa = 3.69QLLNMSEE58 pKa = 4.49EE59 pKa = 4.32LFQFSDD65 pKa = 4.19EE66 pKa = 4.49EE67 pKa = 4.99DD68 pKa = 3.55NSQTPPRR75 pKa = 11.84TSTPEE80 pKa = 3.48QTDD83 pKa = 3.3PKK85 pKa = 10.77VCVDD89 pKa = 3.31NLGIRR94 pKa = 11.84EE95 pKa = 4.31GTGNGTIQLGSEE107 pKa = 4.5SEE109 pKa = 4.25TSLGSVGNSNDD120 pKa = 3.5RR121 pKa = 11.84GKK123 pKa = 10.64KK124 pKa = 6.8RR125 pKa = 11.84QRR127 pKa = 11.84GITYY131 pKa = 10.1ISDD134 pKa = 3.57TSDD137 pKa = 2.95SSGSDD142 pKa = 3.58DD143 pKa = 4.09EE144 pKa = 5.38NLGTPHH150 pKa = 6.87RR151 pKa = 11.84NKK153 pKa = 9.66RR154 pKa = 11.84TRR156 pKa = 11.84NSNTTKK162 pKa = 9.75EE163 pKa = 4.33TGGGDD168 pKa = 3.37NRR170 pKa = 11.84RR171 pKa = 11.84HH172 pKa = 5.48QEE174 pKa = 3.68SDD176 pKa = 2.77HH177 pKa = 6.22GSNGNRR183 pKa = 11.84LEE185 pKa = 4.2PTDD188 pKa = 4.16GGEE191 pKa = 4.31SCSSGTQPDD200 pKa = 5.02FIEE203 pKa = 4.72GTPNGPDD210 pKa = 3.15EE211 pKa = 4.25MDD213 pKa = 3.0GRR215 pKa = 11.84RR216 pKa = 11.84LEE218 pKa = 3.88EE219 pKa = 4.15SEE221 pKa = 3.86IDD223 pKa = 3.57KK224 pKa = 10.85QVEE227 pKa = 4.15STTWYY232 pKa = 9.04TFVIRR237 pKa = 11.84EE238 pKa = 4.04KK239 pKa = 9.67PQPRR243 pKa = 11.84RR244 pKa = 11.84LSGRR248 pKa = 11.84TPNFTITDD256 pKa = 4.0HH257 pKa = 6.73GDD259 pKa = 2.86HH260 pKa = 5.68WHH262 pKa = 6.06ITYY265 pKa = 10.5SGHH268 pKa = 5.99PTNKK272 pKa = 7.59TRR274 pKa = 11.84HH275 pKa = 5.82RR276 pKa = 11.84ATILAYY282 pKa = 10.75LGVTFAARR290 pKa = 11.84AEE292 pKa = 4.22AEE294 pKa = 4.06ATTVLVRR301 pKa = 11.84NIKK304 pKa = 10.03RR305 pKa = 11.84WILYY309 pKa = 8.45LIRR312 pKa = 11.84YY313 pKa = 7.88GIEE316 pKa = 3.49RR317 pKa = 11.84LSYY320 pKa = 10.4FGLGHH325 pKa = 7.53AIFKK329 pKa = 10.55RR330 pKa = 11.84IIKK333 pKa = 9.25YY334 pKa = 7.27FQQYY338 pKa = 9.86RR339 pKa = 11.84RR340 pKa = 11.84DD341 pKa = 3.56EE342 pKa = 4.39DD343 pKa = 4.77AVDD346 pKa = 4.96GPCPYY351 pKa = 8.15MTTTRR356 pKa = 11.84EE357 pKa = 3.77DD358 pKa = 3.41RR359 pKa = 11.84AEE361 pKa = 4.17EE362 pKa = 4.12KK363 pKa = 10.47PKK365 pKa = 10.81EE366 pKa = 3.92NSAEE370 pKa = 3.88YY371 pKa = 10.14DD372 pKa = 3.78YY373 pKa = 11.29LQHH376 pKa = 6.98LVKK379 pKa = 9.86TKK381 pKa = 9.44SARR384 pKa = 11.84TVQEE388 pKa = 4.28LVNKK392 pKa = 10.41LDD394 pKa = 3.88DD395 pKa = 4.09EE396 pKa = 4.77EE397 pKa = 5.64YY398 pKa = 9.93KK399 pKa = 10.68QLWTRR404 pKa = 11.84TRR406 pKa = 11.84GQYY409 pKa = 9.99KK410 pKa = 10.45DD411 pKa = 3.35KK412 pKa = 11.13LRR414 pKa = 11.84GILTYY419 pKa = 11.05YY420 pKa = 10.42NNKK423 pKa = 9.52KK424 pKa = 10.26KK425 pKa = 10.87SNQSQLSLITNLQNISKK442 pKa = 10.41RR443 pKa = 11.84KK444 pKa = 8.29PDD446 pKa = 3.96YY447 pKa = 11.73DD448 pKa = 3.19NMQWIKK454 pKa = 11.46YY455 pKa = 7.86MLANNDD461 pKa = 2.63IRR463 pKa = 11.84VPEE466 pKa = 3.87ILAWIIIVADD476 pKa = 3.85KK477 pKa = 11.04KK478 pKa = 10.74LDD480 pKa = 4.15KK481 pKa = 11.06INTLVLQGPTGTGKK495 pKa = 10.59SLTIGALLGKK505 pKa = 10.23LNTGLVTRR513 pKa = 11.84TGDD516 pKa = 3.32SNTFHH521 pKa = 6.75LQNLIGKK528 pKa = 9.11SYY530 pKa = 11.56ALFEE534 pKa = 4.41EE535 pKa = 4.68PRR537 pKa = 11.84ISQITVDD544 pKa = 4.01DD545 pKa = 4.36FKK547 pKa = 11.82LLFEE551 pKa = 4.99GSDD554 pKa = 3.38LEE556 pKa = 4.9VNIKK560 pKa = 9.77HH561 pKa = 5.57QEE563 pKa = 3.88SEE565 pKa = 3.78IMGRR569 pKa = 11.84IPIFISTNKK578 pKa = 10.59DD579 pKa = 2.13IDD581 pKa = 3.87YY582 pKa = 8.44WVPPADD588 pKa = 3.56GKK590 pKa = 10.96ALQTRR595 pKa = 11.84TKK597 pKa = 9.22TFHH600 pKa = 6.65LTRR603 pKa = 11.84QIKK606 pKa = 10.09GLSDD610 pKa = 3.52RR611 pKa = 11.84MNSQYY616 pKa = 11.04DD617 pKa = 3.63INPPPDD623 pKa = 5.19KK624 pKa = 10.18ITSDD628 pKa = 3.66DD629 pKa = 3.76FLGLFQEE636 pKa = 4.62YY637 pKa = 9.77EE638 pKa = 4.23KK639 pKa = 10.94EE640 pKa = 3.65IDD642 pKa = 4.68DD643 pKa = 5.48IIDD646 pKa = 3.14NHH648 pKa = 5.65VRR650 pKa = 11.84RR651 pKa = 11.84FNKK654 pKa = 10.1SKK656 pKa = 10.22PKK658 pKa = 10.48EE659 pKa = 4.2KK660 pKa = 10.03IQEE663 pKa = 4.24GCTT666 pKa = 3.34
Molecular weight: 75.77 kDa
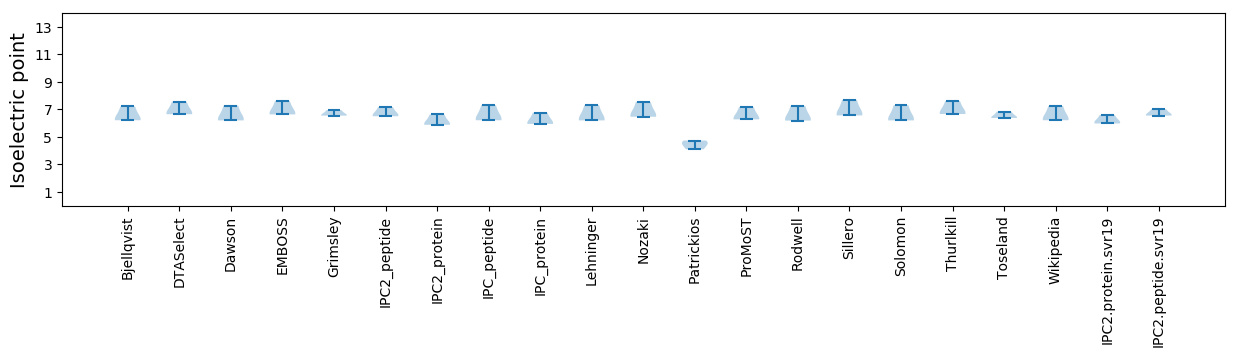
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9IBP6|Q9IBP6_IHHNV Non-structural protein OS=Infectious hypodermal and hematopoietic necrosis virus OX=1513224 PE=4 SV=1
MM1 pKa = 7.45CADD4 pKa = 3.67STRR7 pKa = 11.84ASPRR11 pKa = 11.84KK12 pKa = 9.04RR13 pKa = 11.84SRR15 pKa = 11.84RR16 pKa = 11.84DD17 pKa = 2.94AHH19 pKa = 7.48NEE21 pKa = 3.86DD22 pKa = 4.02EE23 pKa = 4.32EE24 pKa = 4.36HH25 pKa = 7.23AEE27 pKa = 4.38GSSGPDD33 pKa = 3.04PHH35 pKa = 7.45RR36 pKa = 11.84CLQFNTGDD44 pKa = 3.93SIHH47 pKa = 6.5ITFQTRR53 pKa = 11.84RR54 pKa = 11.84YY55 pKa = 10.23FEE57 pKa = 4.23FDD59 pKa = 2.97AANDD63 pKa = 3.83GNFDD67 pKa = 4.09GKK69 pKa = 10.31NLYY72 pKa = 9.9CLPLHH77 pKa = 6.52WMNLYY82 pKa = 10.75LYY84 pKa = 9.21GLKK87 pKa = 10.61SSDD90 pKa = 3.18SSATEE95 pKa = 3.65TQRR98 pKa = 11.84YY99 pKa = 9.58KK100 pKa = 10.15MVKK103 pKa = 10.47SMMKK107 pKa = 7.99TYY109 pKa = 9.6GWKK112 pKa = 8.21VHH114 pKa = 5.67KK115 pKa = 10.34AGVVMHH121 pKa = 6.44SMVPLMKK128 pKa = 10.15DD129 pKa = 3.46LKK131 pKa = 10.83VSGGTSFEE139 pKa = 4.36TLTFTDD145 pKa = 3.65TPYY148 pKa = 11.48LEE150 pKa = 4.37IFKK153 pKa = 9.51DD154 pKa = 3.47TTGLHH159 pKa = 5.71NQLSTKK165 pKa = 10.01EE166 pKa = 3.94ADD168 pKa = 3.15VTLAKK173 pKa = 9.98WIQNPQLVTVQSTAANYY190 pKa = 8.02EE191 pKa = 4.03DD192 pKa = 6.0PIQQFGFMEE201 pKa = 3.91QMRR204 pKa = 11.84TGDD207 pKa = 3.1RR208 pKa = 11.84KK209 pKa = 10.66AYY211 pKa = 8.23TIHH214 pKa = 7.06GDD216 pKa = 3.05TRR218 pKa = 11.84NWYY221 pKa = 9.88GGEE224 pKa = 4.04IPTTGPTFIPKK235 pKa = 9.32WGGQLKK241 pKa = 9.31WDD243 pKa = 4.17KK244 pKa = 10.63PSLGNLVYY252 pKa = 10.1PADD255 pKa = 3.48HH256 pKa = 6.75HH257 pKa = 6.6TNDD260 pKa = 2.99WQQIFMRR267 pKa = 11.84MSPIKK272 pKa = 10.47GPNGDD277 pKa = 4.13EE278 pKa = 4.3LKK280 pKa = 10.86LGCRR284 pKa = 11.84VQADD288 pKa = 4.32FFLHH292 pKa = 6.57LEE294 pKa = 4.12VRR296 pKa = 11.84LPPQGCVASLGMLQYY311 pKa = 11.12LHH313 pKa = 7.0APCTGQLNKK322 pKa = 10.32CYY324 pKa = 10.26IMHH327 pKa = 6.7TNN329 pKa = 3.14
MM1 pKa = 7.45CADD4 pKa = 3.67STRR7 pKa = 11.84ASPRR11 pKa = 11.84KK12 pKa = 9.04RR13 pKa = 11.84SRR15 pKa = 11.84RR16 pKa = 11.84DD17 pKa = 2.94AHH19 pKa = 7.48NEE21 pKa = 3.86DD22 pKa = 4.02EE23 pKa = 4.32EE24 pKa = 4.36HH25 pKa = 7.23AEE27 pKa = 4.38GSSGPDD33 pKa = 3.04PHH35 pKa = 7.45RR36 pKa = 11.84CLQFNTGDD44 pKa = 3.93SIHH47 pKa = 6.5ITFQTRR53 pKa = 11.84RR54 pKa = 11.84YY55 pKa = 10.23FEE57 pKa = 4.23FDD59 pKa = 2.97AANDD63 pKa = 3.83GNFDD67 pKa = 4.09GKK69 pKa = 10.31NLYY72 pKa = 9.9CLPLHH77 pKa = 6.52WMNLYY82 pKa = 10.75LYY84 pKa = 9.21GLKK87 pKa = 10.61SSDD90 pKa = 3.18SSATEE95 pKa = 3.65TQRR98 pKa = 11.84YY99 pKa = 9.58KK100 pKa = 10.15MVKK103 pKa = 10.47SMMKK107 pKa = 7.99TYY109 pKa = 9.6GWKK112 pKa = 8.21VHH114 pKa = 5.67KK115 pKa = 10.34AGVVMHH121 pKa = 6.44SMVPLMKK128 pKa = 10.15DD129 pKa = 3.46LKK131 pKa = 10.83VSGGTSFEE139 pKa = 4.36TLTFTDD145 pKa = 3.65TPYY148 pKa = 11.48LEE150 pKa = 4.37IFKK153 pKa = 9.51DD154 pKa = 3.47TTGLHH159 pKa = 5.71NQLSTKK165 pKa = 10.01EE166 pKa = 3.94ADD168 pKa = 3.15VTLAKK173 pKa = 9.98WIQNPQLVTVQSTAANYY190 pKa = 8.02EE191 pKa = 4.03DD192 pKa = 6.0PIQQFGFMEE201 pKa = 3.91QMRR204 pKa = 11.84TGDD207 pKa = 3.1RR208 pKa = 11.84KK209 pKa = 10.66AYY211 pKa = 8.23TIHH214 pKa = 7.06GDD216 pKa = 3.05TRR218 pKa = 11.84NWYY221 pKa = 9.88GGEE224 pKa = 4.04IPTTGPTFIPKK235 pKa = 9.32WGGQLKK241 pKa = 9.31WDD243 pKa = 4.17KK244 pKa = 10.63PSLGNLVYY252 pKa = 10.1PADD255 pKa = 3.48HH256 pKa = 6.75HH257 pKa = 6.6TNDD260 pKa = 2.99WQQIFMRR267 pKa = 11.84MSPIKK272 pKa = 10.47GPNGDD277 pKa = 4.13EE278 pKa = 4.3LKK280 pKa = 10.86LGCRR284 pKa = 11.84VQADD288 pKa = 4.32FFLHH292 pKa = 6.57LEE294 pKa = 4.12VRR296 pKa = 11.84LPPQGCVASLGMLQYY311 pKa = 11.12LHH313 pKa = 7.0APCTGQLNKK322 pKa = 10.32CYY324 pKa = 10.26IMHH327 pKa = 6.7TNN329 pKa = 3.14
Molecular weight: 37.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1358 |
329 |
666 |
452.7 |
51.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.535 ± 0.581 | 1.105 ± 0.411 |
6.112 ± 0.842 | 7.216 ± 1.054 |
3.019 ± 0.43 | 6.112 ± 1.332 |
2.946 ± 0.493 | 5.817 ± 0.95 |
6.48 ± 0.197 | 8.689 ± 0.523 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.135 ± 0.814 | 5.67 ± 0.386 |
5.744 ± 0.622 | 5.302 ± 0.385 |
6.038 ± 0.652 | 6.48 ± 0.485 |
9.352 ± 0.507 | 3.756 ± 0.521 |
1.694 ± 0.427 | 2.798 ± 0.675 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
