
Rhodopirellula sp. SM50
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Pirellulales; Pirellulaceae; Rhodopirellula; unclassified Rhodopirellula
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
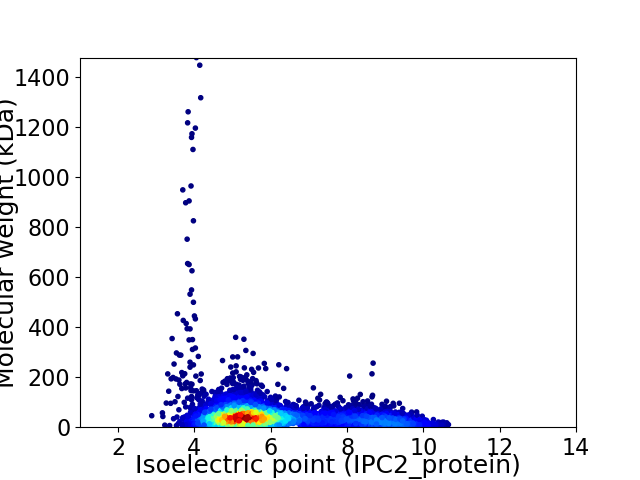
Virtual 2D-PAGE plot for 6414 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2A2W7E3|A0A2A2W7E3_9BACT Peptidase_A24 domain-containing protein OS=Rhodopirellula sp. SM50 OX=595453 GN=CKO51_23740 PE=3 SV=1
MM1 pKa = 7.1NVSRR5 pKa = 11.84QCLFVLFAAGSLCSMAGAEE24 pKa = 4.11TVTFEE29 pKa = 4.66NLLTTPEE36 pKa = 4.05SFYY39 pKa = 11.23KK40 pKa = 10.84GDD42 pKa = 3.66LSQINSDD49 pKa = 3.95PLTVDD54 pKa = 3.02GFEE57 pKa = 4.94FSNQTTYY64 pKa = 11.08VDD66 pKa = 3.94PVTLDD71 pKa = 4.24GYY73 pKa = 9.38WSGWAYY79 pKa = 11.51SNTTDD84 pKa = 2.97TTTPGFANEE93 pKa = 3.91QSAITGGGSNGAGGSVSGEE112 pKa = 4.04TYY114 pKa = 11.22ALAFGSGATINLPAGALIEE133 pKa = 4.5SVDD136 pKa = 3.92WTNGTYY142 pKa = 9.96PYY144 pKa = 11.22LSMRR148 pKa = 11.84DD149 pKa = 2.74GDD151 pKa = 4.36AFAKK155 pKa = 10.28QFGGAGGSEE164 pKa = 3.69PDD166 pKa = 3.73FFRR169 pKa = 11.84VILTGYY175 pKa = 10.31SDD177 pKa = 4.96LNATGTATGSVTLDD191 pKa = 3.19LADD194 pKa = 3.63YY195 pKa = 8.39TFSDD199 pKa = 4.26DD200 pKa = 3.7NQDD203 pKa = 4.03YY204 pKa = 10.84IVDD207 pKa = 3.68AWQVNEE213 pKa = 5.05DD214 pKa = 3.57LTALGNARR222 pKa = 11.84SIDD225 pKa = 3.89LVFQSSDD232 pKa = 2.94SGQFGINTPTYY243 pKa = 10.94LFIDD247 pKa = 3.56NLRR250 pKa = 11.84YY251 pKa = 9.81SVTAIPEE258 pKa = 3.79PTAFGFLALVGGVFTLRR275 pKa = 11.84RR276 pKa = 11.84RR277 pKa = 11.84RR278 pKa = 3.51
MM1 pKa = 7.1NVSRR5 pKa = 11.84QCLFVLFAAGSLCSMAGAEE24 pKa = 4.11TVTFEE29 pKa = 4.66NLLTTPEE36 pKa = 4.05SFYY39 pKa = 11.23KK40 pKa = 10.84GDD42 pKa = 3.66LSQINSDD49 pKa = 3.95PLTVDD54 pKa = 3.02GFEE57 pKa = 4.94FSNQTTYY64 pKa = 11.08VDD66 pKa = 3.94PVTLDD71 pKa = 4.24GYY73 pKa = 9.38WSGWAYY79 pKa = 11.51SNTTDD84 pKa = 2.97TTTPGFANEE93 pKa = 3.91QSAITGGGSNGAGGSVSGEE112 pKa = 4.04TYY114 pKa = 11.22ALAFGSGATINLPAGALIEE133 pKa = 4.5SVDD136 pKa = 3.92WTNGTYY142 pKa = 9.96PYY144 pKa = 11.22LSMRR148 pKa = 11.84DD149 pKa = 2.74GDD151 pKa = 4.36AFAKK155 pKa = 10.28QFGGAGGSEE164 pKa = 3.69PDD166 pKa = 3.73FFRR169 pKa = 11.84VILTGYY175 pKa = 10.31SDD177 pKa = 4.96LNATGTATGSVTLDD191 pKa = 3.19LADD194 pKa = 3.63YY195 pKa = 8.39TFSDD199 pKa = 4.26DD200 pKa = 3.7NQDD203 pKa = 4.03YY204 pKa = 10.84IVDD207 pKa = 3.68AWQVNEE213 pKa = 5.05DD214 pKa = 3.57LTALGNARR222 pKa = 11.84SIDD225 pKa = 3.89LVFQSSDD232 pKa = 2.94SGQFGINTPTYY243 pKa = 10.94LFIDD247 pKa = 3.56NLRR250 pKa = 11.84YY251 pKa = 9.81SVTAIPEE258 pKa = 3.79PTAFGFLALVGGVFTLRR275 pKa = 11.84RR276 pKa = 11.84RR277 pKa = 11.84RR278 pKa = 3.51
Molecular weight: 29.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2A2WIU8|A0A2A2WIU8_9BACT Stage V sporulation protein G OS=Rhodopirellula sp. SM50 OX=595453 GN=CKO51_02225 PE=4 SV=1
MM1 pKa = 7.5RR2 pKa = 11.84CEE4 pKa = 3.64KK5 pKa = 10.38RR6 pKa = 11.84SRR8 pKa = 11.84KK9 pKa = 10.18LGDD12 pKa = 3.31TVGVISRR19 pKa = 11.84WRR21 pKa = 11.84FIRR24 pKa = 11.84TAVKK28 pKa = 10.29APRR31 pKa = 11.84SEE33 pKa = 4.13PDD35 pKa = 3.39TIVFGSQDD43 pKa = 2.85RR44 pKa = 11.84TALMRR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84CLRR54 pKa = 11.84DD55 pKa = 3.39LEE57 pKa = 4.39YY58 pKa = 10.49PPLYY62 pKa = 10.16RR63 pKa = 11.84GEE65 pKa = 4.29VVDD68 pKa = 4.56
MM1 pKa = 7.5RR2 pKa = 11.84CEE4 pKa = 3.64KK5 pKa = 10.38RR6 pKa = 11.84SRR8 pKa = 11.84KK9 pKa = 10.18LGDD12 pKa = 3.31TVGVISRR19 pKa = 11.84WRR21 pKa = 11.84FIRR24 pKa = 11.84TAVKK28 pKa = 10.29APRR31 pKa = 11.84SEE33 pKa = 4.13PDD35 pKa = 3.39TIVFGSQDD43 pKa = 2.85RR44 pKa = 11.84TALMRR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84CLRR54 pKa = 11.84DD55 pKa = 3.39LEE57 pKa = 4.39YY58 pKa = 10.49PPLYY62 pKa = 10.16RR63 pKa = 11.84GEE65 pKa = 4.29VVDD68 pKa = 4.56
Molecular weight: 8.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2788521 |
35 |
13982 |
434.8 |
47.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.856 ± 0.042 | 1.031 ± 0.021 |
6.9 ± 0.053 | 5.816 ± 0.031 |
3.664 ± 0.02 | 7.995 ± 0.056 |
2.15 ± 0.021 | 5.031 ± 0.017 |
3.145 ± 0.039 | 9.529 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.048 ± 0.025 | 3.147 ± 0.033 |
5.214 ± 0.025 | 4.092 ± 0.022 |
6.755 ± 0.051 | 6.752 ± 0.037 |
5.855 ± 0.056 | 7.268 ± 0.03 |
1.431 ± 0.015 | 2.319 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
