
Feline foamy virus (FFV) (Feline syncytial virus)
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; Spumaretrovirinae; Felispumavirus
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
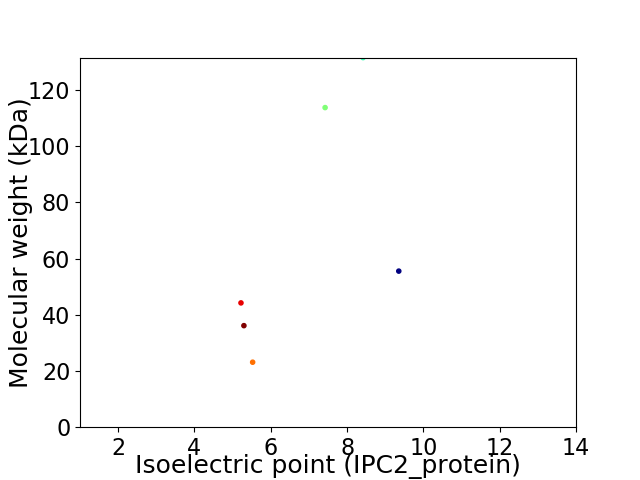
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|O93209|POL_FFV Pro-Pol polyprotein OS=Feline foamy virus OX=53182 GN=pol PE=3 SV=1
MM1 pKa = 7.35ASKK4 pKa = 10.68YY5 pKa = 9.96PEE7 pKa = 4.47EE8 pKa = 4.82GPITEE13 pKa = 4.32GVEE16 pKa = 3.9EE17 pKa = 4.87DD18 pKa = 4.31FNSHH22 pKa = 5.06STSGLDD28 pKa = 3.29LTSVGKK34 pKa = 10.26NPEE37 pKa = 3.76HH38 pKa = 7.09PRR40 pKa = 11.84RR41 pKa = 11.84ILLVLQHH48 pKa = 6.71LIAYY52 pKa = 9.96AEE54 pKa = 4.07ATIKK58 pKa = 10.48KK59 pKa = 9.43QDD61 pKa = 3.46VPGPLLPILSPYY73 pKa = 8.82VMAWDD78 pKa = 3.9NPQNVVTRR86 pKa = 11.84LVNLGEE92 pKa = 4.14SWKK95 pKa = 10.62KK96 pKa = 10.65YY97 pKa = 10.12LLSPGWKK104 pKa = 9.86DD105 pKa = 3.19CGEE108 pKa = 4.43RR109 pKa = 11.84DD110 pKa = 3.29LTMLTRR116 pKa = 11.84EE117 pKa = 4.4LLVPGIGLVQIAATLTKK134 pKa = 10.08TYY136 pKa = 11.53VLMCNGRR143 pKa = 11.84CITGSRR149 pKa = 11.84TDD151 pKa = 4.09PDD153 pKa = 4.59CDD155 pKa = 3.64PLFCKK160 pKa = 10.36LLCWKK165 pKa = 10.5QNIQDD170 pKa = 3.66PRR172 pKa = 11.84EE173 pKa = 4.22CNLEE177 pKa = 3.96EE178 pKa = 3.82WCLYY182 pKa = 10.97SLDD185 pKa = 4.64PEE187 pKa = 5.24HH188 pKa = 7.78DD189 pKa = 4.22PLWDD193 pKa = 3.18PKK195 pKa = 10.5MIVRR199 pKa = 11.84RR200 pKa = 11.84HH201 pKa = 5.87RR202 pKa = 11.84NLLPYY207 pKa = 10.41CMRR210 pKa = 11.84PFLIWMNYY218 pKa = 8.54ISHH221 pKa = 6.89NPLTQQCIMMKK232 pKa = 8.51TLNMLWRR239 pKa = 11.84AQADD243 pKa = 3.93DD244 pKa = 4.37PSDD247 pKa = 3.67VASLYY252 pKa = 10.33PRR254 pKa = 11.84VKK256 pKa = 10.34VFKK259 pKa = 10.34ASHH262 pKa = 6.08FDD264 pKa = 3.25IFGSASGNSEE274 pKa = 4.33EE275 pKa = 4.11RR276 pKa = 11.84VSWAKK281 pKa = 10.44EE282 pKa = 3.45NSHH285 pKa = 6.72RR286 pKa = 11.84GEE288 pKa = 4.06YY289 pKa = 10.47SLLPSSDD296 pKa = 3.93DD297 pKa = 3.62EE298 pKa = 4.32EE299 pKa = 5.66EE300 pKa = 4.2EE301 pKa = 3.88MSEE304 pKa = 4.9RR305 pKa = 11.84EE306 pKa = 3.99EE307 pKa = 4.05LLCHH311 pKa = 7.32INQCQQKK318 pKa = 9.64LFYY321 pKa = 10.34PGGTTDD327 pKa = 3.53VLGMEE332 pKa = 4.57SNVWLTKK339 pKa = 9.95FVNIKK344 pKa = 9.77FPKK347 pKa = 7.17GTKK350 pKa = 9.96VILPDD355 pKa = 3.17GRR357 pKa = 11.84KK358 pKa = 9.46FIACDD363 pKa = 3.68PEE365 pKa = 4.86LKK367 pKa = 10.38PLLQEE372 pKa = 4.94LKK374 pKa = 10.49FLDD377 pKa = 3.98RR378 pKa = 11.84ATSEE382 pKa = 4.33SSDD385 pKa = 3.71SEE387 pKa = 4.27
MM1 pKa = 7.35ASKK4 pKa = 10.68YY5 pKa = 9.96PEE7 pKa = 4.47EE8 pKa = 4.82GPITEE13 pKa = 4.32GVEE16 pKa = 3.9EE17 pKa = 4.87DD18 pKa = 4.31FNSHH22 pKa = 5.06STSGLDD28 pKa = 3.29LTSVGKK34 pKa = 10.26NPEE37 pKa = 3.76HH38 pKa = 7.09PRR40 pKa = 11.84RR41 pKa = 11.84ILLVLQHH48 pKa = 6.71LIAYY52 pKa = 9.96AEE54 pKa = 4.07ATIKK58 pKa = 10.48KK59 pKa = 9.43QDD61 pKa = 3.46VPGPLLPILSPYY73 pKa = 8.82VMAWDD78 pKa = 3.9NPQNVVTRR86 pKa = 11.84LVNLGEE92 pKa = 4.14SWKK95 pKa = 10.62KK96 pKa = 10.65YY97 pKa = 10.12LLSPGWKK104 pKa = 9.86DD105 pKa = 3.19CGEE108 pKa = 4.43RR109 pKa = 11.84DD110 pKa = 3.29LTMLTRR116 pKa = 11.84EE117 pKa = 4.4LLVPGIGLVQIAATLTKK134 pKa = 10.08TYY136 pKa = 11.53VLMCNGRR143 pKa = 11.84CITGSRR149 pKa = 11.84TDD151 pKa = 4.09PDD153 pKa = 4.59CDD155 pKa = 3.64PLFCKK160 pKa = 10.36LLCWKK165 pKa = 10.5QNIQDD170 pKa = 3.66PRR172 pKa = 11.84EE173 pKa = 4.22CNLEE177 pKa = 3.96EE178 pKa = 3.82WCLYY182 pKa = 10.97SLDD185 pKa = 4.64PEE187 pKa = 5.24HH188 pKa = 7.78DD189 pKa = 4.22PLWDD193 pKa = 3.18PKK195 pKa = 10.5MIVRR199 pKa = 11.84RR200 pKa = 11.84HH201 pKa = 5.87RR202 pKa = 11.84NLLPYY207 pKa = 10.41CMRR210 pKa = 11.84PFLIWMNYY218 pKa = 8.54ISHH221 pKa = 6.89NPLTQQCIMMKK232 pKa = 8.51TLNMLWRR239 pKa = 11.84AQADD243 pKa = 3.93DD244 pKa = 4.37PSDD247 pKa = 3.67VASLYY252 pKa = 10.33PRR254 pKa = 11.84VKK256 pKa = 10.34VFKK259 pKa = 10.34ASHH262 pKa = 6.08FDD264 pKa = 3.25IFGSASGNSEE274 pKa = 4.33EE275 pKa = 4.11RR276 pKa = 11.84VSWAKK281 pKa = 10.44EE282 pKa = 3.45NSHH285 pKa = 6.72RR286 pKa = 11.84GEE288 pKa = 4.06YY289 pKa = 10.47SLLPSSDD296 pKa = 3.93DD297 pKa = 3.62EE298 pKa = 4.32EE299 pKa = 5.66EE300 pKa = 4.2EE301 pKa = 3.88MSEE304 pKa = 4.9RR305 pKa = 11.84EE306 pKa = 3.99EE307 pKa = 4.05LLCHH311 pKa = 7.32INQCQQKK318 pKa = 9.64LFYY321 pKa = 10.34PGGTTDD327 pKa = 3.53VLGMEE332 pKa = 4.57SNVWLTKK339 pKa = 9.95FVNIKK344 pKa = 9.77FPKK347 pKa = 7.17GTKK350 pKa = 9.96VILPDD355 pKa = 3.17GRR357 pKa = 11.84KK358 pKa = 9.46FIACDD363 pKa = 3.68PEE365 pKa = 4.86LKK367 pKa = 10.38PLLQEE372 pKa = 4.94LKK374 pKa = 10.49FLDD377 pKa = 3.98RR378 pKa = 11.84ATSEE382 pKa = 4.33SSDD385 pKa = 3.71SEE387 pKa = 4.27
Molecular weight: 44.22 kDa
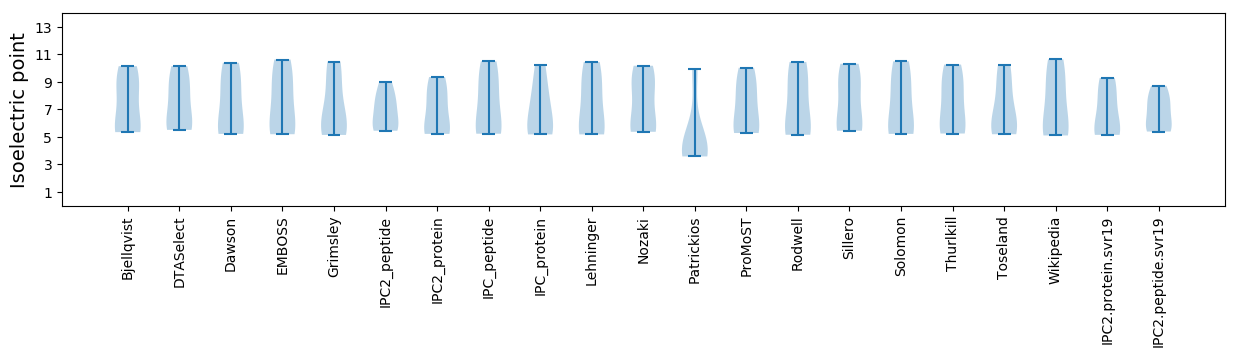
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|O56861|ENV_FFV Envelope glycoprotein gp130 OS=Feline foamy virus OX=53182 GN=env PE=1 SV=1
MM1 pKa = 7.42ARR3 pKa = 11.84EE4 pKa = 4.23LNPLQLQQLYY14 pKa = 10.05INNGLQPNPGHH25 pKa = 6.78GDD27 pKa = 3.61IIAVRR32 pKa = 11.84FTGGPWGPGDD42 pKa = 2.77RR43 pKa = 11.84WARR46 pKa = 11.84VTIRR50 pKa = 11.84LQDD53 pKa = 3.4NTGQPLQVPGYY64 pKa = 9.02DD65 pKa = 4.03LEE67 pKa = 5.24PGIINLRR74 pKa = 11.84EE75 pKa = 4.75DD76 pKa = 3.22ILIAGPYY83 pKa = 9.54NLIRR87 pKa = 11.84TAFLDD92 pKa = 3.77LEE94 pKa = 4.41PARR97 pKa = 11.84GPEE100 pKa = 3.59RR101 pKa = 11.84HH102 pKa = 6.28GPFGDD107 pKa = 3.98GRR109 pKa = 11.84LQPGDD114 pKa = 3.59GLSEE118 pKa = 4.43GFQPITDD125 pKa = 3.89EE126 pKa = 4.63EE127 pKa = 4.24IQAEE131 pKa = 4.39VGTIGAARR139 pKa = 11.84NEE141 pKa = 3.61IRR143 pKa = 11.84LLRR146 pKa = 11.84EE147 pKa = 3.58ALQRR151 pKa = 11.84LQAGGVGRR159 pKa = 11.84PIPGAVLQPQPVIGPVIPINHH180 pKa = 6.27LRR182 pKa = 11.84SVIGNTPPNPRR193 pKa = 11.84DD194 pKa = 3.24VALWLGRR201 pKa = 11.84STAAIEE207 pKa = 4.46GVFPIVDD214 pKa = 3.25QVTRR218 pKa = 11.84MRR220 pKa = 11.84VVNALVASHH229 pKa = 7.41PGLTLTEE236 pKa = 4.26NEE238 pKa = 4.09AGSWNAAISALWRR251 pKa = 11.84KK252 pKa = 8.92AHH254 pKa = 6.16GAAAQHH260 pKa = 5.97EE261 pKa = 4.53LAGVLSDD268 pKa = 3.44INKK271 pKa = 9.9KK272 pKa = 10.51EE273 pKa = 4.34GIQTAFNLGMQFTDD287 pKa = 4.5GNWSLVWGIIRR298 pKa = 11.84TLLPGQALVTNAQSQFDD315 pKa = 4.18LMGDD319 pKa = 4.13DD320 pKa = 3.42IQRR323 pKa = 11.84AEE325 pKa = 4.01NFPRR329 pKa = 11.84VINNLYY335 pKa = 9.96TMLGLNIHH343 pKa = 5.95GQSIRR348 pKa = 11.84PRR350 pKa = 11.84VQTQPLQTRR359 pKa = 11.84PRR361 pKa = 11.84NPGRR365 pKa = 11.84SQQGQLNQPRR375 pKa = 11.84PQNRR379 pKa = 11.84ANQSYY384 pKa = 9.76RR385 pKa = 11.84PPRR388 pKa = 11.84QQQQHH393 pKa = 5.87SDD395 pKa = 3.42VPEE398 pKa = 3.82QRR400 pKa = 11.84DD401 pKa = 3.4QRR403 pKa = 11.84GPSQPPRR410 pKa = 11.84GSGGGYY416 pKa = 8.6NFRR419 pKa = 11.84RR420 pKa = 11.84NPQQPQRR427 pKa = 11.84YY428 pKa = 7.97GQGPPGPNPYY438 pKa = 10.03RR439 pKa = 11.84RR440 pKa = 11.84FGDD443 pKa = 3.53GGNPQQQGPPPNRR456 pKa = 11.84GPDD459 pKa = 3.16QGPRR463 pKa = 11.84PGGNPRR469 pKa = 11.84GGGRR473 pKa = 11.84GQGPRR478 pKa = 11.84NGGGSAAAVHH488 pKa = 5.29TVKK491 pKa = 10.86ASEE494 pKa = 4.19NEE496 pKa = 4.34TKK498 pKa = 10.65NGSAEE503 pKa = 4.0AVDD506 pKa = 3.95GGKK509 pKa = 10.3KK510 pKa = 9.93GGKK513 pKa = 9.22DD514 pKa = 2.99
MM1 pKa = 7.42ARR3 pKa = 11.84EE4 pKa = 4.23LNPLQLQQLYY14 pKa = 10.05INNGLQPNPGHH25 pKa = 6.78GDD27 pKa = 3.61IIAVRR32 pKa = 11.84FTGGPWGPGDD42 pKa = 2.77RR43 pKa = 11.84WARR46 pKa = 11.84VTIRR50 pKa = 11.84LQDD53 pKa = 3.4NTGQPLQVPGYY64 pKa = 9.02DD65 pKa = 4.03LEE67 pKa = 5.24PGIINLRR74 pKa = 11.84EE75 pKa = 4.75DD76 pKa = 3.22ILIAGPYY83 pKa = 9.54NLIRR87 pKa = 11.84TAFLDD92 pKa = 3.77LEE94 pKa = 4.41PARR97 pKa = 11.84GPEE100 pKa = 3.59RR101 pKa = 11.84HH102 pKa = 6.28GPFGDD107 pKa = 3.98GRR109 pKa = 11.84LQPGDD114 pKa = 3.59GLSEE118 pKa = 4.43GFQPITDD125 pKa = 3.89EE126 pKa = 4.63EE127 pKa = 4.24IQAEE131 pKa = 4.39VGTIGAARR139 pKa = 11.84NEE141 pKa = 3.61IRR143 pKa = 11.84LLRR146 pKa = 11.84EE147 pKa = 3.58ALQRR151 pKa = 11.84LQAGGVGRR159 pKa = 11.84PIPGAVLQPQPVIGPVIPINHH180 pKa = 6.27LRR182 pKa = 11.84SVIGNTPPNPRR193 pKa = 11.84DD194 pKa = 3.24VALWLGRR201 pKa = 11.84STAAIEE207 pKa = 4.46GVFPIVDD214 pKa = 3.25QVTRR218 pKa = 11.84MRR220 pKa = 11.84VVNALVASHH229 pKa = 7.41PGLTLTEE236 pKa = 4.26NEE238 pKa = 4.09AGSWNAAISALWRR251 pKa = 11.84KK252 pKa = 8.92AHH254 pKa = 6.16GAAAQHH260 pKa = 5.97EE261 pKa = 4.53LAGVLSDD268 pKa = 3.44INKK271 pKa = 9.9KK272 pKa = 10.51EE273 pKa = 4.34GIQTAFNLGMQFTDD287 pKa = 4.5GNWSLVWGIIRR298 pKa = 11.84TLLPGQALVTNAQSQFDD315 pKa = 4.18LMGDD319 pKa = 4.13DD320 pKa = 3.42IQRR323 pKa = 11.84AEE325 pKa = 4.01NFPRR329 pKa = 11.84VINNLYY335 pKa = 9.96TMLGLNIHH343 pKa = 5.95GQSIRR348 pKa = 11.84PRR350 pKa = 11.84VQTQPLQTRR359 pKa = 11.84PRR361 pKa = 11.84NPGRR365 pKa = 11.84SQQGQLNQPRR375 pKa = 11.84PQNRR379 pKa = 11.84ANQSYY384 pKa = 9.76RR385 pKa = 11.84PPRR388 pKa = 11.84QQQQHH393 pKa = 5.87SDD395 pKa = 3.42VPEE398 pKa = 3.82QRR400 pKa = 11.84DD401 pKa = 3.4QRR403 pKa = 11.84GPSQPPRR410 pKa = 11.84GSGGGYY416 pKa = 8.6NFRR419 pKa = 11.84RR420 pKa = 11.84NPQQPQRR427 pKa = 11.84YY428 pKa = 7.97GQGPPGPNPYY438 pKa = 10.03RR439 pKa = 11.84RR440 pKa = 11.84FGDD443 pKa = 3.53GGNPQQQGPPPNRR456 pKa = 11.84GPDD459 pKa = 3.16QGPRR463 pKa = 11.84PGGNPRR469 pKa = 11.84GGGRR473 pKa = 11.84GQGPRR478 pKa = 11.84NGGGSAAAVHH488 pKa = 5.29TVKK491 pKa = 10.86ASEE494 pKa = 4.19NEE496 pKa = 4.34TKK498 pKa = 10.65NGSAEE503 pKa = 4.0AVDD506 pKa = 3.95GGKK509 pKa = 10.3KK510 pKa = 9.93GGKK513 pKa = 9.22DD514 pKa = 2.99
Molecular weight: 55.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3561 |
209 |
1156 |
593.5 |
67.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.521 ± 0.578 | 1.91 ± 0.613 |
4.802 ± 0.357 | 6.094 ± 0.541 |
3.061 ± 0.249 | 6.712 ± 1.555 |
2.443 ± 0.19 | 6.403 ± 0.718 |
6.375 ± 1.129 | 9.997 ± 0.595 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.825 ± 0.357 | 5.392 ± 0.312 |
6.712 ± 0.807 | 5.336 ± 0.826 |
4.999 ± 0.846 | 6.15 ± 0.634 |
5.981 ± 0.544 | 5.532 ± 0.378 |
2.19 ± 0.224 | 3.566 ± 0.511 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
