
Wenzhou tombus-like virus 4
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.71
Get precalculated fractions of proteins
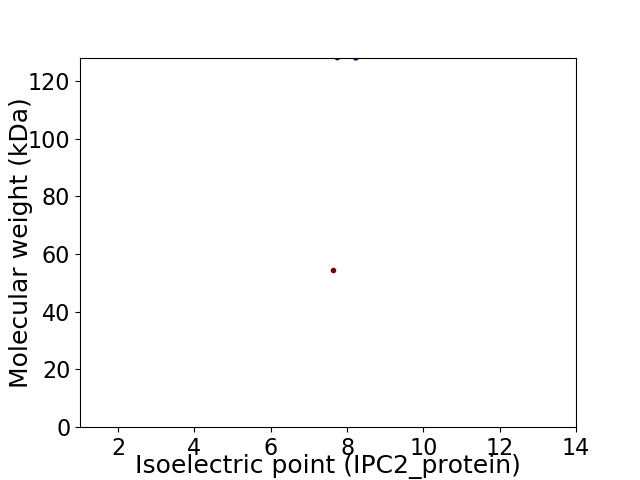
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
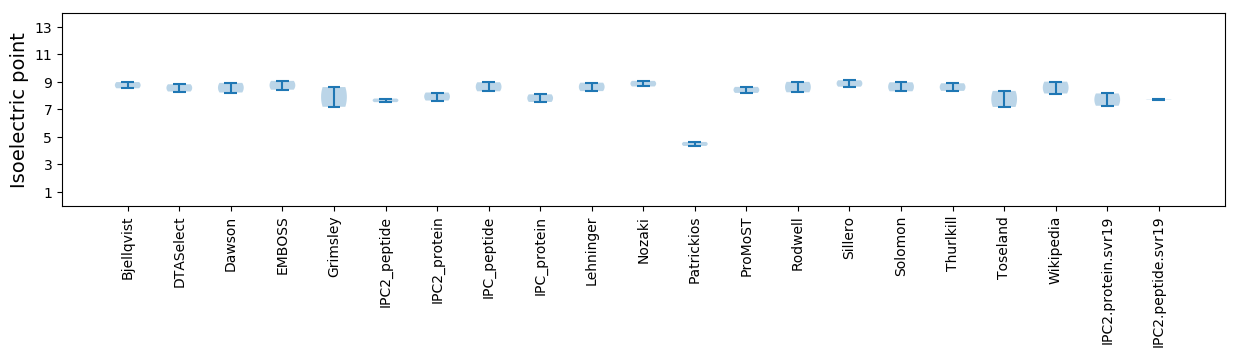
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGC4|A0A1L3KGC4_9VIRU Uncharacterized protein OS=Wenzhou tombus-like virus 4 OX=1923674 PE=4 SV=1
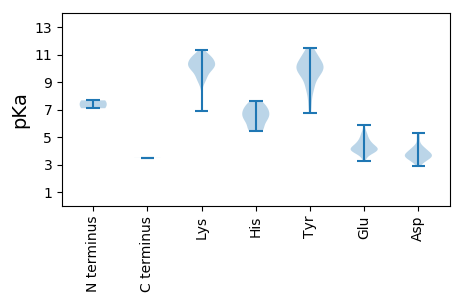
MM1 pKa = 7.1NTTNYY6 pKa = 10.82AGLSDD11 pKa = 5.1LLTAGSRR18 pKa = 11.84SIAYY22 pKa = 9.81SKK24 pKa = 10.82LLSAEE29 pKa = 4.0AAGQLEE35 pKa = 4.71PGAASYY41 pKa = 10.21IISALDD47 pKa = 3.56PFHH50 pKa = 7.59DD51 pKa = 3.94KK52 pKa = 10.96QYY54 pKa = 11.04DD55 pKa = 3.64ATDD58 pKa = 3.62VPSVGVQRR66 pKa = 11.84SIVRR70 pKa = 11.84VLRR73 pKa = 11.84TRR75 pKa = 11.84TRR77 pKa = 11.84ISMEE81 pKa = 3.75SATSVGYY88 pKa = 6.78TQPGRR93 pKa = 11.84LALCMYY99 pKa = 9.75PSVNSRR105 pKa = 11.84YY106 pKa = 8.69MSKK109 pKa = 10.68KK110 pKa = 10.61DD111 pKa = 3.31IVNGQAVDD119 pKa = 3.7PATAAAKK126 pKa = 10.33YY127 pKa = 10.3VGGLVWEE134 pKa = 5.21LDD136 pKa = 3.29IQGQPKK142 pKa = 9.82FFTGDD147 pKa = 3.39PSDD150 pKa = 4.13YY151 pKa = 11.12SADD154 pKa = 3.66ANNGRR159 pKa = 11.84MSIADD164 pKa = 3.6VTGGMPDD171 pKa = 3.43HH172 pKa = 6.64AHH174 pKa = 5.8MRR176 pKa = 11.84VIGCGFEE183 pKa = 3.94VHH185 pKa = 6.25NVTEE189 pKa = 4.29EE190 pKa = 3.92LSKK193 pKa = 11.34GGTVSVWQLPGIFARR208 pKa = 11.84NKK210 pKa = 9.7VVDD213 pKa = 3.82GTQVVTNAPATGIYY227 pKa = 9.43YY228 pKa = 10.56NCDD231 pKa = 3.13FLGRR235 pKa = 11.84APTTADD241 pKa = 3.11EE242 pKa = 4.54VTNIPTGQTWEE253 pKa = 4.22AGQGCLVVPRR263 pKa = 11.84LDD265 pKa = 3.81SGDD268 pKa = 3.85FLLAGCNTPNAEE280 pKa = 4.04ALIEE284 pKa = 4.23GVWDD288 pKa = 3.92GNGTYY293 pKa = 10.2AYY295 pKa = 10.02FPTGSNTSVRR305 pKa = 11.84TFSTPFSMGGALFEE319 pKa = 4.37GLPPEE324 pKa = 4.25SVLDD328 pKa = 4.3VEE330 pKa = 5.3CKK332 pKa = 9.84WIVAEE337 pKa = 4.44VPHH340 pKa = 7.47VSDD343 pKa = 4.71IGDD346 pKa = 3.53LVIARR351 pKa = 11.84NTPPGMFPHH360 pKa = 5.43TTKK363 pKa = 11.13LIACLNRR370 pKa = 11.84TLRR373 pKa = 11.84DD374 pKa = 3.85GYY376 pKa = 9.36PAGWNEE382 pKa = 3.89SGKK385 pKa = 9.96YY386 pKa = 7.15WSSVLRR392 pKa = 11.84AISSVLPMIGNAVPIPGAKK411 pKa = 10.12LLTSAASAALQQKK424 pKa = 9.35ARR426 pKa = 11.84NRR428 pKa = 11.84DD429 pKa = 3.29EE430 pKa = 5.36TIDD433 pKa = 3.74LMKK436 pKa = 10.25DD437 pKa = 3.43LKK439 pKa = 10.55SARR442 pKa = 11.84PGTKK446 pKa = 9.29EE447 pKa = 3.48YY448 pKa = 11.47NEE450 pKa = 5.05LYY452 pKa = 9.59RR453 pKa = 11.84QAMANAGPIQRR464 pKa = 11.84RR465 pKa = 11.84QLRR468 pKa = 11.84RR469 pKa = 11.84LRR471 pKa = 11.84RR472 pKa = 11.84ADD474 pKa = 3.12EE475 pKa = 4.07RR476 pKa = 11.84TVKK479 pKa = 10.23GFTMVSPSTGSKK491 pKa = 9.08TKK493 pKa = 10.85LPTSSIAVVKK503 pKa = 10.58RR504 pKa = 11.84RR505 pKa = 3.48
MM1 pKa = 7.1NTTNYY6 pKa = 10.82AGLSDD11 pKa = 5.1LLTAGSRR18 pKa = 11.84SIAYY22 pKa = 9.81SKK24 pKa = 10.82LLSAEE29 pKa = 4.0AAGQLEE35 pKa = 4.71PGAASYY41 pKa = 10.21IISALDD47 pKa = 3.56PFHH50 pKa = 7.59DD51 pKa = 3.94KK52 pKa = 10.96QYY54 pKa = 11.04DD55 pKa = 3.64ATDD58 pKa = 3.62VPSVGVQRR66 pKa = 11.84SIVRR70 pKa = 11.84VLRR73 pKa = 11.84TRR75 pKa = 11.84TRR77 pKa = 11.84ISMEE81 pKa = 3.75SATSVGYY88 pKa = 6.78TQPGRR93 pKa = 11.84LALCMYY99 pKa = 9.75PSVNSRR105 pKa = 11.84YY106 pKa = 8.69MSKK109 pKa = 10.68KK110 pKa = 10.61DD111 pKa = 3.31IVNGQAVDD119 pKa = 3.7PATAAAKK126 pKa = 10.33YY127 pKa = 10.3VGGLVWEE134 pKa = 5.21LDD136 pKa = 3.29IQGQPKK142 pKa = 9.82FFTGDD147 pKa = 3.39PSDD150 pKa = 4.13YY151 pKa = 11.12SADD154 pKa = 3.66ANNGRR159 pKa = 11.84MSIADD164 pKa = 3.6VTGGMPDD171 pKa = 3.43HH172 pKa = 6.64AHH174 pKa = 5.8MRR176 pKa = 11.84VIGCGFEE183 pKa = 3.94VHH185 pKa = 6.25NVTEE189 pKa = 4.29EE190 pKa = 3.92LSKK193 pKa = 11.34GGTVSVWQLPGIFARR208 pKa = 11.84NKK210 pKa = 9.7VVDD213 pKa = 3.82GTQVVTNAPATGIYY227 pKa = 9.43YY228 pKa = 10.56NCDD231 pKa = 3.13FLGRR235 pKa = 11.84APTTADD241 pKa = 3.11EE242 pKa = 4.54VTNIPTGQTWEE253 pKa = 4.22AGQGCLVVPRR263 pKa = 11.84LDD265 pKa = 3.81SGDD268 pKa = 3.85FLLAGCNTPNAEE280 pKa = 4.04ALIEE284 pKa = 4.23GVWDD288 pKa = 3.92GNGTYY293 pKa = 10.2AYY295 pKa = 10.02FPTGSNTSVRR305 pKa = 11.84TFSTPFSMGGALFEE319 pKa = 4.37GLPPEE324 pKa = 4.25SVLDD328 pKa = 4.3VEE330 pKa = 5.3CKK332 pKa = 9.84WIVAEE337 pKa = 4.44VPHH340 pKa = 7.47VSDD343 pKa = 4.71IGDD346 pKa = 3.53LVIARR351 pKa = 11.84NTPPGMFPHH360 pKa = 5.43TTKK363 pKa = 11.13LIACLNRR370 pKa = 11.84TLRR373 pKa = 11.84DD374 pKa = 3.85GYY376 pKa = 9.36PAGWNEE382 pKa = 3.89SGKK385 pKa = 9.96YY386 pKa = 7.15WSSVLRR392 pKa = 11.84AISSVLPMIGNAVPIPGAKK411 pKa = 10.12LLTSAASAALQQKK424 pKa = 9.35ARR426 pKa = 11.84NRR428 pKa = 11.84DD429 pKa = 3.29EE430 pKa = 5.36TIDD433 pKa = 3.74LMKK436 pKa = 10.25DD437 pKa = 3.43LKK439 pKa = 10.55SARR442 pKa = 11.84PGTKK446 pKa = 9.29EE447 pKa = 3.48YY448 pKa = 11.47NEE450 pKa = 5.05LYY452 pKa = 9.59RR453 pKa = 11.84QAMANAGPIQRR464 pKa = 11.84RR465 pKa = 11.84QLRR468 pKa = 11.84RR469 pKa = 11.84LRR471 pKa = 11.84RR472 pKa = 11.84ADD474 pKa = 3.12EE475 pKa = 4.07RR476 pKa = 11.84TVKK479 pKa = 10.23GFTMVSPSTGSKK491 pKa = 9.08TKK493 pKa = 10.85LPTSSIAVVKK503 pKa = 10.58RR504 pKa = 11.84RR505 pKa = 3.48
Molecular weight: 54.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGC4|A0A1L3KGC4_9VIRU Uncharacterized protein OS=Wenzhou tombus-like virus 4 OX=1923674 PE=4 SV=1
MM1 pKa = 7.67EE2 pKa = 4.21SHH4 pKa = 6.92EE5 pKa = 4.76APGALPEE12 pKa = 4.56GKK14 pKa = 10.09ALPRR18 pKa = 11.84VPADD22 pKa = 3.12NSIYY26 pKa = 9.81MAQLNLAASVLMLLKK41 pKa = 10.8LCFKK45 pKa = 10.47LAYY48 pKa = 9.29AVYY51 pKa = 9.58MFFATIARR59 pKa = 11.84CLIRR63 pKa = 11.84YY64 pKa = 8.68VEE66 pKa = 4.14SLLRR70 pKa = 11.84SEE72 pKa = 5.86LGLPDD77 pKa = 4.81DD78 pKa = 4.49EE79 pKa = 5.5EE80 pKa = 5.13IPFALITGRR89 pKa = 11.84ILKK92 pKa = 9.65LFHH95 pKa = 5.9AAEE98 pKa = 4.15RR99 pKa = 11.84RR100 pKa = 11.84VSAFIQMLGTDD111 pKa = 3.74VPRR114 pKa = 11.84LLEE117 pKa = 4.17EE118 pKa = 5.17LDD120 pKa = 3.62VKK122 pKa = 11.06DD123 pKa = 3.97RR124 pKa = 11.84ATSAGSRR131 pKa = 11.84VVNEE135 pKa = 3.88ASGLLIRR142 pKa = 11.84AAAVVGDD149 pKa = 4.18LRR151 pKa = 11.84DD152 pKa = 3.62LVAEE156 pKa = 4.47EE157 pKa = 4.89LPIPNPANVDD167 pKa = 3.66PGLEE171 pKa = 4.44GYY173 pKa = 10.14VSPEE177 pKa = 3.45LVLNDD182 pKa = 3.69EE183 pKa = 4.8GNLEE187 pKa = 3.89EE188 pKa = 5.0MINHH192 pKa = 6.74EE193 pKa = 4.71LNVAAGKK200 pKa = 9.02MEE202 pKa = 4.43AEE204 pKa = 4.66CVPQEE209 pKa = 4.41HH210 pKa = 6.23TPQTNPVSAINEE222 pKa = 4.2LLQRR226 pKa = 11.84MPVTIHH232 pKa = 5.48TYY234 pKa = 8.19EE235 pKa = 3.85YY236 pKa = 10.32RR237 pKa = 11.84DD238 pKa = 4.1LGVKK242 pKa = 10.1SPTYY246 pKa = 10.25ACTMTLSNVATLKK259 pKa = 10.46EE260 pKa = 4.05GRR262 pKa = 11.84LLQVTAIGGSKK273 pKa = 9.86KK274 pKa = 9.24VAKK277 pKa = 8.91RR278 pKa = 11.84TAAIRR283 pKa = 11.84ACYY286 pKa = 9.82ALGVKK291 pKa = 9.92YY292 pKa = 10.31EE293 pKa = 4.27GMPPTPMKK301 pKa = 10.32TSVKK305 pKa = 9.97LIPIEE310 pKa = 4.1TLGEE314 pKa = 3.81ASYY317 pKa = 11.04KK318 pKa = 10.65KK319 pKa = 10.44YY320 pKa = 10.94VEE322 pKa = 3.82PLKK325 pKa = 11.0FFTADD330 pKa = 3.39EE331 pKa = 4.24NDD333 pKa = 3.71GTGRR337 pKa = 11.84PTMAGPVPKK346 pKa = 10.11AAEE349 pKa = 4.17VLSSMKK355 pKa = 10.21EE356 pKa = 3.85KK357 pKa = 10.26PLKK360 pKa = 10.72LRR362 pKa = 11.84DD363 pKa = 3.76AKK365 pKa = 10.66SVSMTVKK372 pKa = 10.35QLIEE376 pKa = 3.64DD377 pKa = 4.44TYY379 pKa = 10.97RR380 pKa = 11.84VLVRR384 pKa = 11.84EE385 pKa = 4.08GLKK388 pKa = 8.97PTLAQLLEE396 pKa = 4.09RR397 pKa = 11.84VARR400 pKa = 11.84NNPGPVAEE408 pKa = 4.31VVGVVQEE415 pKa = 4.17MEE417 pKa = 4.44EE418 pKa = 4.2VADD421 pKa = 3.93THH423 pKa = 6.35SHH425 pKa = 6.37AGLPTVVPVPAAPMPVVYY443 pKa = 10.41DD444 pKa = 3.38PTGPDD449 pKa = 3.36GEE451 pKa = 4.68YY452 pKa = 11.0EE453 pKa = 4.12PMDD456 pKa = 4.13PPGDD460 pKa = 4.09PGEE463 pKa = 5.29DD464 pKa = 4.18DD465 pKa = 5.03PPPDD469 pKa = 4.3PPGGPDD475 pKa = 3.96GDD477 pKa = 4.33RR478 pKa = 11.84LPADD482 pKa = 4.87LDD484 pKa = 3.81PSVVDD489 pKa = 3.17TVVALVRR496 pKa = 11.84RR497 pKa = 11.84NHH499 pKa = 6.73PSPPRR504 pKa = 11.84NDD506 pKa = 4.22AEE508 pKa = 4.18AQSQRR513 pKa = 11.84NAANAEE519 pKa = 3.64LDD521 pKa = 3.75AYY523 pKa = 10.25FEE525 pKa = 4.52TMMADD530 pKa = 3.6GTVHH534 pKa = 6.47ARR536 pKa = 11.84KK537 pKa = 10.02LMAARR542 pKa = 11.84YY543 pKa = 8.57IDD545 pKa = 3.7PVVTKK550 pKa = 9.96LEE552 pKa = 4.11HH553 pKa = 5.64VAPSRR558 pKa = 11.84AEE560 pKa = 3.74HH561 pKa = 6.13LLVRR565 pKa = 11.84NAPSVTAKK573 pKa = 10.35RR574 pKa = 11.84VLEE577 pKa = 4.0RR578 pKa = 11.84LGSPGKK584 pKa = 6.9XWCRR588 pKa = 11.84GALRR592 pKa = 11.84GVRR595 pKa = 11.84MLAEE599 pKa = 4.52PDD601 pKa = 2.99WRR603 pKa = 11.84SVNRR607 pKa = 11.84QLPRR611 pKa = 11.84GYY613 pKa = 11.05LLLFKK618 pKa = 10.26PLYY621 pKa = 10.45GGLASRR627 pKa = 11.84RR628 pKa = 11.84RR629 pKa = 11.84LTQLVPGLSLLDD641 pKa = 3.93FCVHH645 pKa = 7.35DD646 pKa = 3.84PTAYY650 pKa = 10.24NVLDD654 pKa = 3.9GVIQRR659 pKa = 11.84VMLVDD664 pKa = 3.29GHH666 pKa = 6.71RR667 pKa = 11.84PPQPEE672 pKa = 3.84PGAWAALRR680 pKa = 11.84RR681 pKa = 11.84YY682 pKa = 9.37ARR684 pKa = 11.84RR685 pKa = 11.84ITRR688 pKa = 11.84RR689 pKa = 11.84MPRR692 pKa = 11.84NPKK695 pKa = 9.45RR696 pKa = 11.84MRR698 pKa = 11.84RR699 pKa = 11.84HH700 pKa = 7.28DD701 pKa = 3.72YY702 pKa = 8.4PQLYY706 pKa = 9.94GGSKK710 pKa = 9.48RR711 pKa = 11.84DD712 pKa = 3.49MYY714 pKa = 11.3AAAAVSLDD722 pKa = 3.7EE723 pKa = 5.05KK724 pKa = 10.76PITSRR729 pKa = 11.84DD730 pKa = 2.93ARR732 pKa = 11.84LAVFGKK738 pKa = 9.82CEE740 pKa = 3.91KK741 pKa = 10.58VALAANGRR749 pKa = 11.84PKK751 pKa = 10.92AMRR754 pKa = 11.84VISPRR759 pKa = 11.84DD760 pKa = 3.14PRR762 pKa = 11.84FNIEE766 pKa = 4.09LGRR769 pKa = 11.84FLHH772 pKa = 6.67PIEE775 pKa = 4.82KK776 pKa = 9.61PVYY779 pKa = 9.13LAINAMFSNTPTTPTVQKK797 pKa = 10.33FLNLRR802 pKa = 11.84EE803 pKa = 3.98RR804 pKa = 11.84GRR806 pKa = 11.84VLRR809 pKa = 11.84AKK811 pKa = 9.35WRR813 pKa = 11.84MFRR816 pKa = 11.84DD817 pKa = 5.21PIAVGYY823 pKa = 8.62DD824 pKa = 2.99AKK826 pKa = 10.95RR827 pKa = 11.84FDD829 pKa = 3.51QHH831 pKa = 5.5TSADD835 pKa = 3.63AMKK838 pKa = 10.16LEE840 pKa = 3.93HH841 pKa = 7.15LIYY844 pKa = 10.03TGAYY848 pKa = 8.74ANHH851 pKa = 7.46PEE853 pKa = 3.96LSEE856 pKa = 4.87LRR858 pKa = 11.84RR859 pKa = 11.84LLNWQLRR866 pKa = 11.84TKK868 pKa = 9.79ATARR872 pKa = 11.84LKK874 pKa = 10.93DD875 pKa = 3.49GLIHH879 pKa = 6.98YY880 pKa = 7.57RR881 pKa = 11.84TLGNRR886 pKa = 11.84CSGDD890 pKa = 3.43VNTSLGNIVLMCVMMRR906 pKa = 11.84AYY908 pKa = 10.47LDD910 pKa = 3.56HH911 pKa = 6.27VRR913 pKa = 11.84LRR915 pKa = 11.84AEE917 pKa = 4.32LVNDD921 pKa = 3.98GDD923 pKa = 4.18DD924 pKa = 3.43CVLILEE930 pKa = 4.59RR931 pKa = 11.84RR932 pKa = 11.84DD933 pKa = 3.41LALLEE938 pKa = 4.04EE939 pKa = 4.85LEE941 pKa = 4.43PFFLRR946 pKa = 11.84LGYY949 pKa = 9.44TMIRR953 pKa = 11.84EE954 pKa = 4.2DD955 pKa = 3.68PVDD958 pKa = 3.45VFEE961 pKa = 5.66QIEE964 pKa = 4.57FCQCNPVFDD973 pKa = 3.96GAEE976 pKa = 3.44WVMVRR981 pKa = 11.84KK982 pKa = 8.54LTTVMSKK989 pKa = 9.3DD990 pKa = 3.44TITVKK995 pKa = 10.68SLSSEE1000 pKa = 3.84QAVRR1004 pKa = 11.84IHH1006 pKa = 6.97RR1007 pKa = 11.84RR1008 pKa = 11.84RR1009 pKa = 11.84IMQSGMAMAGNIPVLRR1025 pKa = 11.84SVYY1028 pKa = 10.17KK1029 pKa = 10.37YY1030 pKa = 10.05IGRR1033 pKa = 11.84GCEE1036 pKa = 3.28AATNRR1041 pKa = 11.84RR1042 pKa = 11.84GRR1044 pKa = 11.84FNPYY1048 pKa = 9.62LDD1050 pKa = 3.95SGLFYY1055 pKa = 9.88ATKK1058 pKa = 10.13GMRR1061 pKa = 11.84YY1062 pKa = 9.91KK1063 pKa = 10.9DD1064 pKa = 3.58ILPTATARR1072 pKa = 11.84LSFWRR1077 pKa = 11.84STGVTPSQQVSMEE1090 pKa = 4.1KK1091 pKa = 10.33QLDD1094 pKa = 4.09DD1095 pKa = 5.33LPADD1099 pKa = 4.23PPDD1102 pKa = 3.41MGLINHH1108 pKa = 7.07RR1109 pKa = 11.84FAAMMFARR1117 pKa = 11.84HH1118 pKa = 5.43NAKK1121 pKa = 10.05PPNLTGSDD1129 pKa = 4.03PLIGYY1134 pKa = 7.73YY1135 pKa = 10.52DD1136 pKa = 4.4RR1137 pKa = 11.84DD1138 pKa = 3.72WPGPAAYY1145 pKa = 9.68RR1146 pKa = 11.84RR1147 pKa = 11.84RR1148 pKa = 11.84QAA1150 pKa = 3.48
MM1 pKa = 7.67EE2 pKa = 4.21SHH4 pKa = 6.92EE5 pKa = 4.76APGALPEE12 pKa = 4.56GKK14 pKa = 10.09ALPRR18 pKa = 11.84VPADD22 pKa = 3.12NSIYY26 pKa = 9.81MAQLNLAASVLMLLKK41 pKa = 10.8LCFKK45 pKa = 10.47LAYY48 pKa = 9.29AVYY51 pKa = 9.58MFFATIARR59 pKa = 11.84CLIRR63 pKa = 11.84YY64 pKa = 8.68VEE66 pKa = 4.14SLLRR70 pKa = 11.84SEE72 pKa = 5.86LGLPDD77 pKa = 4.81DD78 pKa = 4.49EE79 pKa = 5.5EE80 pKa = 5.13IPFALITGRR89 pKa = 11.84ILKK92 pKa = 9.65LFHH95 pKa = 5.9AAEE98 pKa = 4.15RR99 pKa = 11.84RR100 pKa = 11.84VSAFIQMLGTDD111 pKa = 3.74VPRR114 pKa = 11.84LLEE117 pKa = 4.17EE118 pKa = 5.17LDD120 pKa = 3.62VKK122 pKa = 11.06DD123 pKa = 3.97RR124 pKa = 11.84ATSAGSRR131 pKa = 11.84VVNEE135 pKa = 3.88ASGLLIRR142 pKa = 11.84AAAVVGDD149 pKa = 4.18LRR151 pKa = 11.84DD152 pKa = 3.62LVAEE156 pKa = 4.47EE157 pKa = 4.89LPIPNPANVDD167 pKa = 3.66PGLEE171 pKa = 4.44GYY173 pKa = 10.14VSPEE177 pKa = 3.45LVLNDD182 pKa = 3.69EE183 pKa = 4.8GNLEE187 pKa = 3.89EE188 pKa = 5.0MINHH192 pKa = 6.74EE193 pKa = 4.71LNVAAGKK200 pKa = 9.02MEE202 pKa = 4.43AEE204 pKa = 4.66CVPQEE209 pKa = 4.41HH210 pKa = 6.23TPQTNPVSAINEE222 pKa = 4.2LLQRR226 pKa = 11.84MPVTIHH232 pKa = 5.48TYY234 pKa = 8.19EE235 pKa = 3.85YY236 pKa = 10.32RR237 pKa = 11.84DD238 pKa = 4.1LGVKK242 pKa = 10.1SPTYY246 pKa = 10.25ACTMTLSNVATLKK259 pKa = 10.46EE260 pKa = 4.05GRR262 pKa = 11.84LLQVTAIGGSKK273 pKa = 9.86KK274 pKa = 9.24VAKK277 pKa = 8.91RR278 pKa = 11.84TAAIRR283 pKa = 11.84ACYY286 pKa = 9.82ALGVKK291 pKa = 9.92YY292 pKa = 10.31EE293 pKa = 4.27GMPPTPMKK301 pKa = 10.32TSVKK305 pKa = 9.97LIPIEE310 pKa = 4.1TLGEE314 pKa = 3.81ASYY317 pKa = 11.04KK318 pKa = 10.65KK319 pKa = 10.44YY320 pKa = 10.94VEE322 pKa = 3.82PLKK325 pKa = 11.0FFTADD330 pKa = 3.39EE331 pKa = 4.24NDD333 pKa = 3.71GTGRR337 pKa = 11.84PTMAGPVPKK346 pKa = 10.11AAEE349 pKa = 4.17VLSSMKK355 pKa = 10.21EE356 pKa = 3.85KK357 pKa = 10.26PLKK360 pKa = 10.72LRR362 pKa = 11.84DD363 pKa = 3.76AKK365 pKa = 10.66SVSMTVKK372 pKa = 10.35QLIEE376 pKa = 3.64DD377 pKa = 4.44TYY379 pKa = 10.97RR380 pKa = 11.84VLVRR384 pKa = 11.84EE385 pKa = 4.08GLKK388 pKa = 8.97PTLAQLLEE396 pKa = 4.09RR397 pKa = 11.84VARR400 pKa = 11.84NNPGPVAEE408 pKa = 4.31VVGVVQEE415 pKa = 4.17MEE417 pKa = 4.44EE418 pKa = 4.2VADD421 pKa = 3.93THH423 pKa = 6.35SHH425 pKa = 6.37AGLPTVVPVPAAPMPVVYY443 pKa = 10.41DD444 pKa = 3.38PTGPDD449 pKa = 3.36GEE451 pKa = 4.68YY452 pKa = 11.0EE453 pKa = 4.12PMDD456 pKa = 4.13PPGDD460 pKa = 4.09PGEE463 pKa = 5.29DD464 pKa = 4.18DD465 pKa = 5.03PPPDD469 pKa = 4.3PPGGPDD475 pKa = 3.96GDD477 pKa = 4.33RR478 pKa = 11.84LPADD482 pKa = 4.87LDD484 pKa = 3.81PSVVDD489 pKa = 3.17TVVALVRR496 pKa = 11.84RR497 pKa = 11.84NHH499 pKa = 6.73PSPPRR504 pKa = 11.84NDD506 pKa = 4.22AEE508 pKa = 4.18AQSQRR513 pKa = 11.84NAANAEE519 pKa = 3.64LDD521 pKa = 3.75AYY523 pKa = 10.25FEE525 pKa = 4.52TMMADD530 pKa = 3.6GTVHH534 pKa = 6.47ARR536 pKa = 11.84KK537 pKa = 10.02LMAARR542 pKa = 11.84YY543 pKa = 8.57IDD545 pKa = 3.7PVVTKK550 pKa = 9.96LEE552 pKa = 4.11HH553 pKa = 5.64VAPSRR558 pKa = 11.84AEE560 pKa = 3.74HH561 pKa = 6.13LLVRR565 pKa = 11.84NAPSVTAKK573 pKa = 10.35RR574 pKa = 11.84VLEE577 pKa = 4.0RR578 pKa = 11.84LGSPGKK584 pKa = 6.9XWCRR588 pKa = 11.84GALRR592 pKa = 11.84GVRR595 pKa = 11.84MLAEE599 pKa = 4.52PDD601 pKa = 2.99WRR603 pKa = 11.84SVNRR607 pKa = 11.84QLPRR611 pKa = 11.84GYY613 pKa = 11.05LLLFKK618 pKa = 10.26PLYY621 pKa = 10.45GGLASRR627 pKa = 11.84RR628 pKa = 11.84RR629 pKa = 11.84LTQLVPGLSLLDD641 pKa = 3.93FCVHH645 pKa = 7.35DD646 pKa = 3.84PTAYY650 pKa = 10.24NVLDD654 pKa = 3.9GVIQRR659 pKa = 11.84VMLVDD664 pKa = 3.29GHH666 pKa = 6.71RR667 pKa = 11.84PPQPEE672 pKa = 3.84PGAWAALRR680 pKa = 11.84RR681 pKa = 11.84YY682 pKa = 9.37ARR684 pKa = 11.84RR685 pKa = 11.84ITRR688 pKa = 11.84RR689 pKa = 11.84MPRR692 pKa = 11.84NPKK695 pKa = 9.45RR696 pKa = 11.84MRR698 pKa = 11.84RR699 pKa = 11.84HH700 pKa = 7.28DD701 pKa = 3.72YY702 pKa = 8.4PQLYY706 pKa = 9.94GGSKK710 pKa = 9.48RR711 pKa = 11.84DD712 pKa = 3.49MYY714 pKa = 11.3AAAAVSLDD722 pKa = 3.7EE723 pKa = 5.05KK724 pKa = 10.76PITSRR729 pKa = 11.84DD730 pKa = 2.93ARR732 pKa = 11.84LAVFGKK738 pKa = 9.82CEE740 pKa = 3.91KK741 pKa = 10.58VALAANGRR749 pKa = 11.84PKK751 pKa = 10.92AMRR754 pKa = 11.84VISPRR759 pKa = 11.84DD760 pKa = 3.14PRR762 pKa = 11.84FNIEE766 pKa = 4.09LGRR769 pKa = 11.84FLHH772 pKa = 6.67PIEE775 pKa = 4.82KK776 pKa = 9.61PVYY779 pKa = 9.13LAINAMFSNTPTTPTVQKK797 pKa = 10.33FLNLRR802 pKa = 11.84EE803 pKa = 3.98RR804 pKa = 11.84GRR806 pKa = 11.84VLRR809 pKa = 11.84AKK811 pKa = 9.35WRR813 pKa = 11.84MFRR816 pKa = 11.84DD817 pKa = 5.21PIAVGYY823 pKa = 8.62DD824 pKa = 2.99AKK826 pKa = 10.95RR827 pKa = 11.84FDD829 pKa = 3.51QHH831 pKa = 5.5TSADD835 pKa = 3.63AMKK838 pKa = 10.16LEE840 pKa = 3.93HH841 pKa = 7.15LIYY844 pKa = 10.03TGAYY848 pKa = 8.74ANHH851 pKa = 7.46PEE853 pKa = 3.96LSEE856 pKa = 4.87LRR858 pKa = 11.84RR859 pKa = 11.84LLNWQLRR866 pKa = 11.84TKK868 pKa = 9.79ATARR872 pKa = 11.84LKK874 pKa = 10.93DD875 pKa = 3.49GLIHH879 pKa = 6.98YY880 pKa = 7.57RR881 pKa = 11.84TLGNRR886 pKa = 11.84CSGDD890 pKa = 3.43VNTSLGNIVLMCVMMRR906 pKa = 11.84AYY908 pKa = 10.47LDD910 pKa = 3.56HH911 pKa = 6.27VRR913 pKa = 11.84LRR915 pKa = 11.84AEE917 pKa = 4.32LVNDD921 pKa = 3.98GDD923 pKa = 4.18DD924 pKa = 3.43CVLILEE930 pKa = 4.59RR931 pKa = 11.84RR932 pKa = 11.84DD933 pKa = 3.41LALLEE938 pKa = 4.04EE939 pKa = 4.85LEE941 pKa = 4.43PFFLRR946 pKa = 11.84LGYY949 pKa = 9.44TMIRR953 pKa = 11.84EE954 pKa = 4.2DD955 pKa = 3.68PVDD958 pKa = 3.45VFEE961 pKa = 5.66QIEE964 pKa = 4.57FCQCNPVFDD973 pKa = 3.96GAEE976 pKa = 3.44WVMVRR981 pKa = 11.84KK982 pKa = 8.54LTTVMSKK989 pKa = 9.3DD990 pKa = 3.44TITVKK995 pKa = 10.68SLSSEE1000 pKa = 3.84QAVRR1004 pKa = 11.84IHH1006 pKa = 6.97RR1007 pKa = 11.84RR1008 pKa = 11.84RR1009 pKa = 11.84IMQSGMAMAGNIPVLRR1025 pKa = 11.84SVYY1028 pKa = 10.17KK1029 pKa = 10.37YY1030 pKa = 10.05IGRR1033 pKa = 11.84GCEE1036 pKa = 3.28AATNRR1041 pKa = 11.84RR1042 pKa = 11.84GRR1044 pKa = 11.84FNPYY1048 pKa = 9.62LDD1050 pKa = 3.95SGLFYY1055 pKa = 9.88ATKK1058 pKa = 10.13GMRR1061 pKa = 11.84YY1062 pKa = 9.91KK1063 pKa = 10.9DD1064 pKa = 3.58ILPTATARR1072 pKa = 11.84LSFWRR1077 pKa = 11.84STGVTPSQQVSMEE1090 pKa = 4.1KK1091 pKa = 10.33QLDD1094 pKa = 4.09DD1095 pKa = 5.33LPADD1099 pKa = 4.23PPDD1102 pKa = 3.41MGLINHH1108 pKa = 7.07RR1109 pKa = 11.84FAAMMFARR1117 pKa = 11.84HH1118 pKa = 5.43NAKK1121 pKa = 10.05PPNLTGSDD1129 pKa = 4.03PLIGYY1134 pKa = 7.73YY1135 pKa = 10.52DD1136 pKa = 4.4RR1137 pKa = 11.84DD1138 pKa = 3.72WPGPAAYY1145 pKa = 9.68RR1146 pKa = 11.84RR1147 pKa = 11.84RR1148 pKa = 11.84QAA1150 pKa = 3.48
Molecular weight: 127.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1655 |
505 |
1150 |
827.5 |
91.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.849 ± 0.026 | 1.269 ± 0.06 |
5.619 ± 0.139 | 5.438 ± 0.752 |
2.477 ± 0.049 | 7.13 ± 1.007 |
1.752 ± 0.287 | 3.807 ± 0.381 |
4.23 ± 0.137 | 9.728 ± 1.121 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.384 ± 0.412 | 3.807 ± 0.28 |
7.432 ± 0.658 | 2.598 ± 0.29 |
8.036 ± 0.966 | 5.498 ± 1.233 |
5.801 ± 0.978 | 7.795 ± 0.037 |
0.906 ± 0.244 | 3.384 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
