
Roseomonas sp. CQN31
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Acetobacteraceae; Roseomonas
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins
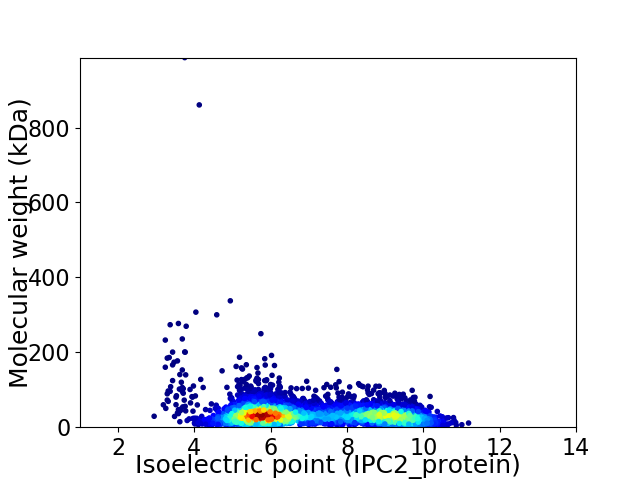
Virtual 2D-PAGE plot for 5423 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A317FD27|A0A317FD27_9PROT Uncharacterized protein OS=Roseomonas sp. CQN31 OX=2184016 GN=DFH01_20465 PE=3 SV=1
MM1 pKa = 7.16ATVSINFGDD10 pKa = 4.75DD11 pKa = 2.93NDD13 pKa = 3.87NFYY16 pKa = 10.65GQPYY20 pKa = 10.42DD21 pKa = 4.42DD22 pKa = 5.63DD23 pKa = 6.4LIIQMGKK30 pKa = 10.87GNDD33 pKa = 3.27TVFAGEE39 pKa = 4.25GTNLVYY45 pKa = 10.75LGAGNDD51 pKa = 3.57VFEE54 pKa = 5.51SEE56 pKa = 5.16DD57 pKa = 3.49MLGGPFDD64 pKa = 4.29EE65 pKa = 4.9STEE68 pKa = 4.35CGPSTVYY75 pKa = 11.22GDD77 pKa = 3.44IGNDD81 pKa = 2.99LFDD84 pKa = 5.37FYY86 pKa = 11.36DD87 pKa = 3.66VSGTAYY93 pKa = 10.18GGKK96 pKa = 10.34DD97 pKa = 2.94NDD99 pKa = 3.34IYY101 pKa = 10.88LIKK104 pKa = 10.91GMDD107 pKa = 3.25ASVSVVEE114 pKa = 4.43YY115 pKa = 10.23GGEE118 pKa = 4.32GIDD121 pKa = 4.5TIKK124 pKa = 10.45IDD126 pKa = 3.73LWSKK130 pKa = 9.02ATYY133 pKa = 7.96TIPVNVEE140 pKa = 3.6NLSVGNVYY148 pKa = 10.65YY149 pKa = 10.2WGFFSNTWEE158 pKa = 4.07TTNNSFEE165 pKa = 4.33EE166 pKa = 4.94GFVDD170 pKa = 4.83AGVTIIGNAEE180 pKa = 3.69NNAITGSDD188 pKa = 3.1MHH190 pKa = 6.77DD191 pKa = 3.58TLKK194 pKa = 11.13GGAGADD200 pKa = 3.68TVKK203 pKa = 10.91GGLANDD209 pKa = 3.7VLHH212 pKa = 7.03GEE214 pKa = 4.78DD215 pKa = 4.56GNDD218 pKa = 3.69VLWGEE223 pKa = 4.4HH224 pKa = 6.42GNDD227 pKa = 3.41KK228 pKa = 11.01AYY230 pKa = 10.76GGIGNDD236 pKa = 3.42KK237 pKa = 10.97LSGHH241 pKa = 6.72GGNDD245 pKa = 3.31TLDD248 pKa = 3.59GGEE251 pKa = 4.49NDD253 pKa = 4.68DD254 pKa = 5.23LVDD257 pKa = 4.01GGGGNDD263 pKa = 4.26LLYY266 pKa = 10.99GGNGKK271 pKa = 9.64DD272 pKa = 3.56VLFDD276 pKa = 3.47GAGADD281 pKa = 3.86TMNGGAGDD289 pKa = 4.92DD290 pKa = 3.72IYY292 pKa = 11.75VVTDD296 pKa = 3.3ATDD299 pKa = 3.5KK300 pKa = 11.07VIEE303 pKa = 4.21AAGGGNDD310 pKa = 4.08LVKK313 pKa = 10.19TKK315 pKa = 10.38LASYY319 pKa = 8.93TLGSEE324 pKa = 4.43VEE326 pKa = 4.34SLMFTGSGGFQGTGNALTNRR346 pKa = 11.84ITGGSGADD354 pKa = 3.74LLKK357 pKa = 11.29GMDD360 pKa = 4.09GNDD363 pKa = 3.07VLTGNDD369 pKa = 4.23GADD372 pKa = 3.26QLYY375 pKa = 9.48TGTGNDD381 pKa = 3.61YY382 pKa = 11.37ASGGNGIDD390 pKa = 3.61SLFGEE395 pKa = 4.96AGNDD399 pKa = 3.23SLYY402 pKa = 11.18GNNDD406 pKa = 2.97ADD408 pKa = 4.12YY409 pKa = 9.24LTGGGDD415 pKa = 3.61NDD417 pKa = 4.18YY418 pKa = 11.47LDD420 pKa = 4.89GGAGNDD426 pKa = 3.95NMHH429 pKa = 7.08GDD431 pKa = 4.24DD432 pKa = 5.12GHH434 pKa = 7.04DD435 pKa = 3.35TMQGGDD441 pKa = 4.36GNDD444 pKa = 3.48LMQGGAGNDD453 pKa = 3.76EE454 pKa = 3.91IRR456 pKa = 11.84GGNGDD461 pKa = 3.75DD462 pKa = 4.58KK463 pKa = 11.24IYY465 pKa = 11.22GGLGKK470 pKa = 10.34DD471 pKa = 3.71KK472 pKa = 10.93LYY474 pKa = 11.34GDD476 pKa = 4.53AGFDD480 pKa = 3.47SFSYY484 pKa = 10.72QSLNEE489 pKa = 3.97SNAAFGRR496 pKa = 11.84DD497 pKa = 3.98FIYY500 pKa = 10.42GFEE503 pKa = 3.97QGKK506 pKa = 9.96DD507 pKa = 3.38RR508 pKa = 11.84IDD510 pKa = 4.08LSWIDD515 pKa = 3.79ADD517 pKa = 3.86STVAGNQGFNFTGSGAFFKK536 pKa = 10.98SAGDD540 pKa = 3.41LWLTSSLLGATVNCDD555 pKa = 3.24VNGDD559 pKa = 3.82AVADD563 pKa = 3.74MQINVVGVWNLTASDD578 pKa = 4.18FVLL581 pKa = 4.61
MM1 pKa = 7.16ATVSINFGDD10 pKa = 4.75DD11 pKa = 2.93NDD13 pKa = 3.87NFYY16 pKa = 10.65GQPYY20 pKa = 10.42DD21 pKa = 4.42DD22 pKa = 5.63DD23 pKa = 6.4LIIQMGKK30 pKa = 10.87GNDD33 pKa = 3.27TVFAGEE39 pKa = 4.25GTNLVYY45 pKa = 10.75LGAGNDD51 pKa = 3.57VFEE54 pKa = 5.51SEE56 pKa = 5.16DD57 pKa = 3.49MLGGPFDD64 pKa = 4.29EE65 pKa = 4.9STEE68 pKa = 4.35CGPSTVYY75 pKa = 11.22GDD77 pKa = 3.44IGNDD81 pKa = 2.99LFDD84 pKa = 5.37FYY86 pKa = 11.36DD87 pKa = 3.66VSGTAYY93 pKa = 10.18GGKK96 pKa = 10.34DD97 pKa = 2.94NDD99 pKa = 3.34IYY101 pKa = 10.88LIKK104 pKa = 10.91GMDD107 pKa = 3.25ASVSVVEE114 pKa = 4.43YY115 pKa = 10.23GGEE118 pKa = 4.32GIDD121 pKa = 4.5TIKK124 pKa = 10.45IDD126 pKa = 3.73LWSKK130 pKa = 9.02ATYY133 pKa = 7.96TIPVNVEE140 pKa = 3.6NLSVGNVYY148 pKa = 10.65YY149 pKa = 10.2WGFFSNTWEE158 pKa = 4.07TTNNSFEE165 pKa = 4.33EE166 pKa = 4.94GFVDD170 pKa = 4.83AGVTIIGNAEE180 pKa = 3.69NNAITGSDD188 pKa = 3.1MHH190 pKa = 6.77DD191 pKa = 3.58TLKK194 pKa = 11.13GGAGADD200 pKa = 3.68TVKK203 pKa = 10.91GGLANDD209 pKa = 3.7VLHH212 pKa = 7.03GEE214 pKa = 4.78DD215 pKa = 4.56GNDD218 pKa = 3.69VLWGEE223 pKa = 4.4HH224 pKa = 6.42GNDD227 pKa = 3.41KK228 pKa = 11.01AYY230 pKa = 10.76GGIGNDD236 pKa = 3.42KK237 pKa = 10.97LSGHH241 pKa = 6.72GGNDD245 pKa = 3.31TLDD248 pKa = 3.59GGEE251 pKa = 4.49NDD253 pKa = 4.68DD254 pKa = 5.23LVDD257 pKa = 4.01GGGGNDD263 pKa = 4.26LLYY266 pKa = 10.99GGNGKK271 pKa = 9.64DD272 pKa = 3.56VLFDD276 pKa = 3.47GAGADD281 pKa = 3.86TMNGGAGDD289 pKa = 4.92DD290 pKa = 3.72IYY292 pKa = 11.75VVTDD296 pKa = 3.3ATDD299 pKa = 3.5KK300 pKa = 11.07VIEE303 pKa = 4.21AAGGGNDD310 pKa = 4.08LVKK313 pKa = 10.19TKK315 pKa = 10.38LASYY319 pKa = 8.93TLGSEE324 pKa = 4.43VEE326 pKa = 4.34SLMFTGSGGFQGTGNALTNRR346 pKa = 11.84ITGGSGADD354 pKa = 3.74LLKK357 pKa = 11.29GMDD360 pKa = 4.09GNDD363 pKa = 3.07VLTGNDD369 pKa = 4.23GADD372 pKa = 3.26QLYY375 pKa = 9.48TGTGNDD381 pKa = 3.61YY382 pKa = 11.37ASGGNGIDD390 pKa = 3.61SLFGEE395 pKa = 4.96AGNDD399 pKa = 3.23SLYY402 pKa = 11.18GNNDD406 pKa = 2.97ADD408 pKa = 4.12YY409 pKa = 9.24LTGGGDD415 pKa = 3.61NDD417 pKa = 4.18YY418 pKa = 11.47LDD420 pKa = 4.89GGAGNDD426 pKa = 3.95NMHH429 pKa = 7.08GDD431 pKa = 4.24DD432 pKa = 5.12GHH434 pKa = 7.04DD435 pKa = 3.35TMQGGDD441 pKa = 4.36GNDD444 pKa = 3.48LMQGGAGNDD453 pKa = 3.76EE454 pKa = 3.91IRR456 pKa = 11.84GGNGDD461 pKa = 3.75DD462 pKa = 4.58KK463 pKa = 11.24IYY465 pKa = 11.22GGLGKK470 pKa = 10.34DD471 pKa = 3.71KK472 pKa = 10.93LYY474 pKa = 11.34GDD476 pKa = 4.53AGFDD480 pKa = 3.47SFSYY484 pKa = 10.72QSLNEE489 pKa = 3.97SNAAFGRR496 pKa = 11.84DD497 pKa = 3.98FIYY500 pKa = 10.42GFEE503 pKa = 3.97QGKK506 pKa = 9.96DD507 pKa = 3.38RR508 pKa = 11.84IDD510 pKa = 4.08LSWIDD515 pKa = 3.79ADD517 pKa = 3.86STVAGNQGFNFTGSGAFFKK536 pKa = 10.98SAGDD540 pKa = 3.41LWLTSSLLGATVNCDD555 pKa = 3.24VNGDD559 pKa = 3.82AVADD563 pKa = 3.74MQINVVGVWNLTASDD578 pKa = 4.18FVLL581 pKa = 4.61
Molecular weight: 60.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A317FHE3|A0A317FHE3_9PROT HTH marR-type domain-containing protein OS=Roseomonas sp. CQN31 OX=2184016 GN=DFH01_02640 PE=4 SV=1
MM1 pKa = 7.56PRR3 pKa = 11.84ACSAARR9 pKa = 11.84TSWKK13 pKa = 9.88PSPPPSRR20 pKa = 11.84RR21 pKa = 11.84SARR24 pKa = 11.84RR25 pKa = 11.84PPSRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84PSPPRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84WWPRR45 pKa = 11.84SRR47 pKa = 11.84KK48 pKa = 8.97SPRR51 pKa = 11.84AARR54 pKa = 11.84TPSSHH59 pKa = 3.92WRR61 pKa = 11.84KK62 pKa = 9.39RR63 pKa = 11.84ATPPPTRR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84APRR75 pKa = 11.84WMARR79 pKa = 11.84RR80 pKa = 11.84SSCARR85 pKa = 11.84SASSGRR91 pKa = 11.84WRR93 pKa = 3.76
MM1 pKa = 7.56PRR3 pKa = 11.84ACSAARR9 pKa = 11.84TSWKK13 pKa = 9.88PSPPPSRR20 pKa = 11.84RR21 pKa = 11.84SARR24 pKa = 11.84RR25 pKa = 11.84PPSRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84PSPPRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84WWPRR45 pKa = 11.84SRR47 pKa = 11.84KK48 pKa = 8.97SPRR51 pKa = 11.84AARR54 pKa = 11.84TPSSHH59 pKa = 3.92WRR61 pKa = 11.84KK62 pKa = 9.39RR63 pKa = 11.84ATPPPTRR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84APRR75 pKa = 11.84WMARR79 pKa = 11.84RR80 pKa = 11.84SSCARR85 pKa = 11.84SASSGRR91 pKa = 11.84WRR93 pKa = 3.76
Molecular weight: 11.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1823058 |
26 |
9467 |
336.2 |
35.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.265 ± 0.068 | 0.817 ± 0.011 |
5.213 ± 0.055 | 5.431 ± 0.034 |
3.361 ± 0.021 | 9.676 ± 0.06 |
1.929 ± 0.02 | 4.026 ± 0.027 |
1.841 ± 0.026 | 10.823 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.297 ± 0.021 | 2.052 ± 0.024 |
6.162 ± 0.04 | 2.845 ± 0.019 |
8.348 ± 0.051 | 4.251 ± 0.027 |
4.99 ± 0.043 | 7.499 ± 0.029 |
1.507 ± 0.015 | 1.667 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
