
Perch rhabdovirus (isolate Perch/France/PRV/1981)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Perhabdovirus; Egli virus
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
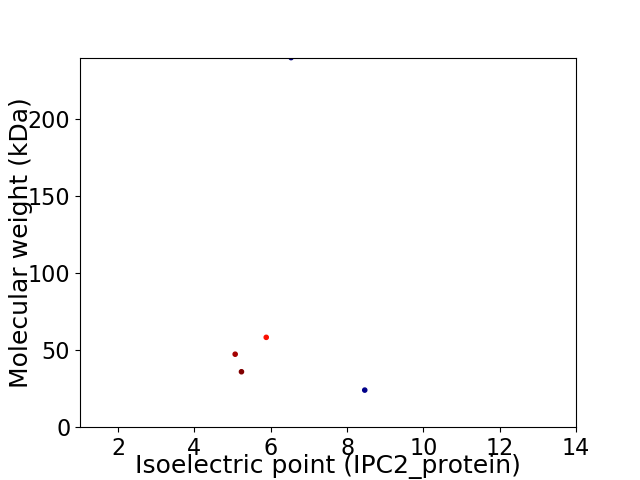
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K7X6R3|K7X6R3_PRVF Matrix protein OS=Perch rhabdovirus (isolate Perch/France/PRV/1981) OX=1559359 GN=M PE=4 SV=1
MM1 pKa = 7.93DD2 pKa = 5.15AGAIVVRR9 pKa = 11.84STKK12 pKa = 10.68ASFVPLQPQLEE23 pKa = 4.27DD24 pKa = 4.16EE25 pKa = 4.49INYY28 pKa = 9.24PADD31 pKa = 3.34FFVNQKK37 pKa = 9.32IPTFVRR43 pKa = 11.84YY44 pKa = 9.43YY45 pKa = 10.87EE46 pKa = 4.01NAKK49 pKa = 10.68PEE51 pKa = 4.54DD52 pKa = 3.99LLGMIFGEE60 pKa = 4.38LSEE63 pKa = 4.5SRR65 pKa = 11.84LPSEE69 pKa = 5.05LVTSYY74 pKa = 10.33IYY76 pKa = 11.15SVMSKK81 pKa = 7.22WTEE84 pKa = 3.82TLDD87 pKa = 4.43APWEE91 pKa = 4.22SFGIEE96 pKa = 3.94FGDD99 pKa = 3.86VNDD102 pKa = 3.68ILTPFSLLKK111 pKa = 10.83CIVSDD116 pKa = 4.89QALADD121 pKa = 3.98YY122 pKa = 8.23KK123 pKa = 10.48TVAVPGDD130 pKa = 3.65IDD132 pKa = 5.05PIAMMIFLLAPYY144 pKa = 9.64RR145 pKa = 11.84IVGIQNEE152 pKa = 4.61EE153 pKa = 3.78YY154 pKa = 9.16QARR157 pKa = 11.84VIQTIQNQIDD167 pKa = 3.66SLGAKK172 pKa = 9.37RR173 pKa = 11.84LNVRR177 pKa = 11.84SLKK180 pKa = 10.89NITTLTRR187 pKa = 11.84SPNYY191 pKa = 9.72LKK193 pKa = 10.81LVAAIDD199 pKa = 3.28MFYY202 pKa = 11.09FHH204 pKa = 7.49FKK206 pKa = 10.44NSQEE210 pKa = 4.06RR211 pKa = 11.84AVVRR215 pKa = 11.84VATLGSRR222 pKa = 11.84HH223 pKa = 6.47KK224 pKa = 10.91DD225 pKa = 3.16CAALSTLNHH234 pKa = 6.08IVQFTGKK241 pKa = 9.39PLIEE245 pKa = 4.01VLDD248 pKa = 3.82WVFTDD253 pKa = 3.94QVAQEE258 pKa = 4.51VSRR261 pKa = 11.84MMRR264 pKa = 11.84PGQEE268 pKa = 3.22IDD270 pKa = 3.38QADD273 pKa = 3.8SYY275 pKa = 11.27MPYY278 pKa = 10.71LKK280 pKa = 10.68DD281 pKa = 4.11LGLCRR286 pKa = 11.84KK287 pKa = 9.59SPYY290 pKa = 9.96SSSANPGVHH299 pKa = 6.14CWAQMTCALLGSKK312 pKa = 10.24RR313 pKa = 11.84SQNAIASTEE322 pKa = 4.0EE323 pKa = 3.69NLVNLTRR330 pKa = 11.84NAEE333 pKa = 3.59IMAYY337 pKa = 10.19VLGTGAVLVKK347 pKa = 10.28ALEE350 pKa = 4.1IGGVKK355 pKa = 9.26GTDD358 pKa = 3.37PDD360 pKa = 5.57DD361 pKa = 3.37ITVDD365 pKa = 3.06VDD367 pKa = 3.82GAGEE371 pKa = 4.15PTTLEE376 pKa = 4.0ALDD379 pKa = 3.75WLDD382 pKa = 4.09FVQTNGCQLTPKK394 pKa = 9.12MEE396 pKa = 4.16AKK398 pKa = 9.84IRR400 pKa = 11.84VMSLRR405 pKa = 11.84IQNARR410 pKa = 11.84KK411 pKa = 8.73DD412 pKa = 3.73TIGAYY417 pKa = 10.33LKK419 pKa = 10.82GRR421 pKa = 11.84AVQNEE426 pKa = 3.91
MM1 pKa = 7.93DD2 pKa = 5.15AGAIVVRR9 pKa = 11.84STKK12 pKa = 10.68ASFVPLQPQLEE23 pKa = 4.27DD24 pKa = 4.16EE25 pKa = 4.49INYY28 pKa = 9.24PADD31 pKa = 3.34FFVNQKK37 pKa = 9.32IPTFVRR43 pKa = 11.84YY44 pKa = 9.43YY45 pKa = 10.87EE46 pKa = 4.01NAKK49 pKa = 10.68PEE51 pKa = 4.54DD52 pKa = 3.99LLGMIFGEE60 pKa = 4.38LSEE63 pKa = 4.5SRR65 pKa = 11.84LPSEE69 pKa = 5.05LVTSYY74 pKa = 10.33IYY76 pKa = 11.15SVMSKK81 pKa = 7.22WTEE84 pKa = 3.82TLDD87 pKa = 4.43APWEE91 pKa = 4.22SFGIEE96 pKa = 3.94FGDD99 pKa = 3.86VNDD102 pKa = 3.68ILTPFSLLKK111 pKa = 10.83CIVSDD116 pKa = 4.89QALADD121 pKa = 3.98YY122 pKa = 8.23KK123 pKa = 10.48TVAVPGDD130 pKa = 3.65IDD132 pKa = 5.05PIAMMIFLLAPYY144 pKa = 9.64RR145 pKa = 11.84IVGIQNEE152 pKa = 4.61EE153 pKa = 3.78YY154 pKa = 9.16QARR157 pKa = 11.84VIQTIQNQIDD167 pKa = 3.66SLGAKK172 pKa = 9.37RR173 pKa = 11.84LNVRR177 pKa = 11.84SLKK180 pKa = 10.89NITTLTRR187 pKa = 11.84SPNYY191 pKa = 9.72LKK193 pKa = 10.81LVAAIDD199 pKa = 3.28MFYY202 pKa = 11.09FHH204 pKa = 7.49FKK206 pKa = 10.44NSQEE210 pKa = 4.06RR211 pKa = 11.84AVVRR215 pKa = 11.84VATLGSRR222 pKa = 11.84HH223 pKa = 6.47KK224 pKa = 10.91DD225 pKa = 3.16CAALSTLNHH234 pKa = 6.08IVQFTGKK241 pKa = 9.39PLIEE245 pKa = 4.01VLDD248 pKa = 3.82WVFTDD253 pKa = 3.94QVAQEE258 pKa = 4.51VSRR261 pKa = 11.84MMRR264 pKa = 11.84PGQEE268 pKa = 3.22IDD270 pKa = 3.38QADD273 pKa = 3.8SYY275 pKa = 11.27MPYY278 pKa = 10.71LKK280 pKa = 10.68DD281 pKa = 4.11LGLCRR286 pKa = 11.84KK287 pKa = 9.59SPYY290 pKa = 9.96SSSANPGVHH299 pKa = 6.14CWAQMTCALLGSKK312 pKa = 10.24RR313 pKa = 11.84SQNAIASTEE322 pKa = 4.0EE323 pKa = 3.69NLVNLTRR330 pKa = 11.84NAEE333 pKa = 3.59IMAYY337 pKa = 10.19VLGTGAVLVKK347 pKa = 10.28ALEE350 pKa = 4.1IGGVKK355 pKa = 9.26GTDD358 pKa = 3.37PDD360 pKa = 5.57DD361 pKa = 3.37ITVDD365 pKa = 3.06VDD367 pKa = 3.82GAGEE371 pKa = 4.15PTTLEE376 pKa = 4.0ALDD379 pKa = 3.75WLDD382 pKa = 4.09FVQTNGCQLTPKK394 pKa = 9.12MEE396 pKa = 4.16AKK398 pKa = 9.84IRR400 pKa = 11.84VMSLRR405 pKa = 11.84IQNARR410 pKa = 11.84KK411 pKa = 8.73DD412 pKa = 3.73TIGAYY417 pKa = 10.33LKK419 pKa = 10.82GRR421 pKa = 11.84AVQNEE426 pKa = 3.91
Molecular weight: 47.39 kDa
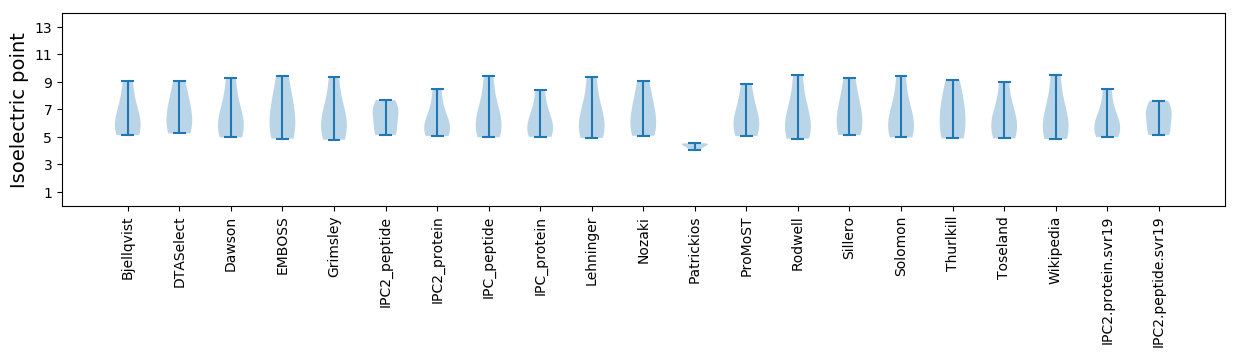
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K7X7F6|K7X7F6_PRVF GDP polyribonucleotidyltransferase OS=Perch rhabdovirus (isolate Perch/France/PRV/1981) OX=1559359 GN=L PE=4 SV=1
MM1 pKa = 7.71ALNIFKK7 pKa = 10.82KK8 pKa = 10.06KK9 pKa = 10.24GKK11 pKa = 9.4SNKK14 pKa = 9.77EE15 pKa = 3.66MDD17 pKa = 3.61SGKK20 pKa = 10.63ALAMPSAPPVYY31 pKa = 10.31SEE33 pKa = 4.85APGPFDD39 pKa = 3.27AWNTSSIEE47 pKa = 4.03EE48 pKa = 4.12LSKK51 pKa = 11.08VSYY54 pKa = 10.78LVDD57 pKa = 3.19TCLSVTTKK65 pKa = 10.94GPLQSVADD73 pKa = 4.41AYY75 pKa = 10.33MVAQGMMDD83 pKa = 4.12YY84 pKa = 11.32YY85 pKa = 11.06NGPVLSKK92 pKa = 10.36PFFMTLFLGGVHH104 pKa = 6.77GMQASVRR111 pKa = 11.84GSQSIRR117 pKa = 11.84YY118 pKa = 7.89EE119 pKa = 4.23RR120 pKa = 11.84EE121 pKa = 3.23FHH123 pKa = 7.06GPMTFPYY130 pKa = 9.82HH131 pKa = 6.93KK132 pKa = 10.17SNPVDD137 pKa = 3.37WTPKK141 pKa = 9.6SVSVHH146 pKa = 4.49YY147 pKa = 9.69TSSLRR152 pKa = 11.84GKK154 pKa = 9.17AVEE157 pKa = 4.35VVFEE161 pKa = 4.43ARR163 pKa = 11.84LKK165 pKa = 9.6STRR168 pKa = 11.84QMGPSVWHH176 pKa = 6.09YY177 pKa = 11.64LDD179 pKa = 3.68GLKK182 pKa = 10.73GIQRR186 pKa = 11.84PGNEE190 pKa = 3.68AVLKK194 pKa = 10.1QFGVPTMMLGNNLVFNLDD212 pKa = 3.34VQMSDD217 pKa = 3.22LL218 pKa = 4.06
MM1 pKa = 7.71ALNIFKK7 pKa = 10.82KK8 pKa = 10.06KK9 pKa = 10.24GKK11 pKa = 9.4SNKK14 pKa = 9.77EE15 pKa = 3.66MDD17 pKa = 3.61SGKK20 pKa = 10.63ALAMPSAPPVYY31 pKa = 10.31SEE33 pKa = 4.85APGPFDD39 pKa = 3.27AWNTSSIEE47 pKa = 4.03EE48 pKa = 4.12LSKK51 pKa = 11.08VSYY54 pKa = 10.78LVDD57 pKa = 3.19TCLSVTTKK65 pKa = 10.94GPLQSVADD73 pKa = 4.41AYY75 pKa = 10.33MVAQGMMDD83 pKa = 4.12YY84 pKa = 11.32YY85 pKa = 11.06NGPVLSKK92 pKa = 10.36PFFMTLFLGGVHH104 pKa = 6.77GMQASVRR111 pKa = 11.84GSQSIRR117 pKa = 11.84YY118 pKa = 7.89EE119 pKa = 4.23RR120 pKa = 11.84EE121 pKa = 3.23FHH123 pKa = 7.06GPMTFPYY130 pKa = 9.82HH131 pKa = 6.93KK132 pKa = 10.17SNPVDD137 pKa = 3.37WTPKK141 pKa = 9.6SVSVHH146 pKa = 4.49YY147 pKa = 9.69TSSLRR152 pKa = 11.84GKK154 pKa = 9.17AVEE157 pKa = 4.35VVFEE161 pKa = 4.43ARR163 pKa = 11.84LKK165 pKa = 9.6STRR168 pKa = 11.84QMGPSVWHH176 pKa = 6.09YY177 pKa = 11.64LDD179 pKa = 3.68GLKK182 pKa = 10.73GIQRR186 pKa = 11.84PGNEE190 pKa = 3.68AVLKK194 pKa = 10.1QFGVPTMMLGNNLVFNLDD212 pKa = 3.34VQMSDD217 pKa = 3.22LL218 pKa = 4.06
Molecular weight: 24.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3579 |
218 |
2097 |
715.8 |
81.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.393 ± 0.525 | 1.369 ± 0.3 |
5.644 ± 0.237 | 6.901 ± 0.45 |
4.135 ± 0.107 | 6.315 ± 0.461 |
2.431 ± 0.297 | 6.175 ± 0.599 |
6.566 ± 0.433 | 9.5 ± 0.604 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.822 ± 0.425 | 4.582 ± 0.171 |
4.806 ± 0.847 | 3.688 ± 0.231 |
5.225 ± 0.516 | 7.907 ± 0.357 |
5.728 ± 0.303 | 5.868 ± 0.805 |
2.012 ± 0.168 | 2.934 ± 0.268 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
