
Rhododendron virus A
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Amalgaviridae; Amalgavirus
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
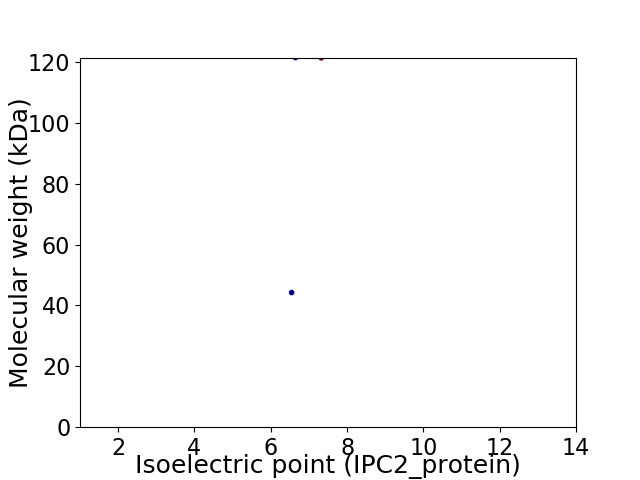
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E1B261|E1B261_9VIRU Putative coat protein OS=Rhododendron virus A OX=878260 PE=4 SV=1
MM1 pKa = 7.78EE2 pKa = 5.6DD3 pKa = 3.06RR4 pKa = 11.84RR5 pKa = 11.84AEE7 pKa = 4.13DD8 pKa = 4.46RR9 pKa = 11.84LDD11 pKa = 4.38DD12 pKa = 4.86DD13 pKa = 5.18LPAGGGAPPPLRR25 pKa = 11.84GLPPLPRR32 pKa = 11.84AEE34 pKa = 4.14PTDD37 pKa = 4.25AEE39 pKa = 4.13LQAAIDD45 pKa = 3.66EE46 pKa = 4.61AVAVVYY52 pKa = 9.38EE53 pKa = 4.19AGMPAGRR60 pKa = 11.84FTVQRR65 pKa = 11.84VHH67 pKa = 7.07EE68 pKa = 4.6IGLTVEE74 pKa = 4.45VFVKK78 pKa = 9.63QARR81 pKa = 11.84AVFGGKK87 pKa = 9.29NVDD90 pKa = 3.42QADD93 pKa = 4.34LIFTTGIKK101 pKa = 9.67MGVCGSLRR109 pKa = 11.84TMEE112 pKa = 4.42PAGFWEE118 pKa = 4.71VIRR121 pKa = 11.84WARR124 pKa = 11.84SNVGRR129 pKa = 11.84QALEE133 pKa = 3.77TGQKK137 pKa = 8.59VKK139 pKa = 10.69KK140 pKa = 10.82VEE142 pKa = 4.21DD143 pKa = 3.63KK144 pKa = 11.06RR145 pKa = 11.84AGTQTPNEE153 pKa = 4.1VALCQIFTMQQGIMAEE169 pKa = 4.18EE170 pKa = 4.24VKK172 pKa = 10.32EE173 pKa = 4.08ARR175 pKa = 11.84ASTQQEE181 pKa = 3.64IDD183 pKa = 3.19EE184 pKa = 4.33LTRR187 pKa = 11.84LLRR190 pKa = 11.84LKK192 pKa = 10.34RR193 pKa = 11.84AEE195 pKa = 4.0QVKK198 pKa = 10.22VLAEE202 pKa = 4.01IKK204 pKa = 10.48DD205 pKa = 3.61KK206 pKa = 11.31YY207 pKa = 10.69FPANIWEE214 pKa = 4.41EE215 pKa = 3.79PAEE218 pKa = 4.08AEE220 pKa = 4.08RR221 pKa = 11.84NARR224 pKa = 11.84CWEE227 pKa = 4.25VYY229 pKa = 10.07SAALVTAGRR238 pKa = 11.84PAPIKK243 pKa = 10.02TEE245 pKa = 3.85AAFKK249 pKa = 10.6LAIDD253 pKa = 4.29AYY255 pKa = 9.86KK256 pKa = 11.02NFVDD260 pKa = 4.13TEE262 pKa = 4.18FKK264 pKa = 10.52TRR266 pKa = 11.84FIRR269 pKa = 11.84SDD271 pKa = 3.14EE272 pKa = 4.18HH273 pKa = 7.42QVALRR278 pKa = 11.84KK279 pKa = 9.17FADD282 pKa = 3.64EE283 pKa = 5.12RR284 pKa = 11.84IHH286 pKa = 6.66FLDD289 pKa = 6.09GIGEE293 pKa = 4.17PKK295 pKa = 10.29KK296 pKa = 10.81AGTFRR301 pKa = 11.84SLLAVLGVEE310 pKa = 4.99PSAQTTTHH318 pKa = 5.1TEE320 pKa = 3.69EE321 pKa = 5.61RR322 pKa = 11.84NDD324 pKa = 3.55EE325 pKa = 4.29DD326 pKa = 5.44DD327 pKa = 4.46SGGEE331 pKa = 3.8SDD333 pKa = 3.7VARR336 pKa = 11.84GEE338 pKa = 4.16EE339 pKa = 4.32PEE341 pKa = 4.34PEE343 pKa = 4.38AGGGTSRR350 pKa = 11.84PGASGRR356 pKa = 11.84GAGRR360 pKa = 11.84GKK362 pKa = 10.52GKK364 pKa = 10.25RR365 pKa = 11.84KK366 pKa = 9.41VVPEE370 pKa = 3.84RR371 pKa = 11.84RR372 pKa = 11.84SLRR375 pKa = 11.84SSKK378 pKa = 9.29TGRR381 pKa = 11.84VTDD384 pKa = 3.53VSVQGAQGDD393 pKa = 4.3GGASRR398 pKa = 11.84PKK400 pKa = 9.9HH401 pKa = 5.49RR402 pKa = 11.84RR403 pKa = 11.84RR404 pKa = 3.44
MM1 pKa = 7.78EE2 pKa = 5.6DD3 pKa = 3.06RR4 pKa = 11.84RR5 pKa = 11.84AEE7 pKa = 4.13DD8 pKa = 4.46RR9 pKa = 11.84LDD11 pKa = 4.38DD12 pKa = 4.86DD13 pKa = 5.18LPAGGGAPPPLRR25 pKa = 11.84GLPPLPRR32 pKa = 11.84AEE34 pKa = 4.14PTDD37 pKa = 4.25AEE39 pKa = 4.13LQAAIDD45 pKa = 3.66EE46 pKa = 4.61AVAVVYY52 pKa = 9.38EE53 pKa = 4.19AGMPAGRR60 pKa = 11.84FTVQRR65 pKa = 11.84VHH67 pKa = 7.07EE68 pKa = 4.6IGLTVEE74 pKa = 4.45VFVKK78 pKa = 9.63QARR81 pKa = 11.84AVFGGKK87 pKa = 9.29NVDD90 pKa = 3.42QADD93 pKa = 4.34LIFTTGIKK101 pKa = 9.67MGVCGSLRR109 pKa = 11.84TMEE112 pKa = 4.42PAGFWEE118 pKa = 4.71VIRR121 pKa = 11.84WARR124 pKa = 11.84SNVGRR129 pKa = 11.84QALEE133 pKa = 3.77TGQKK137 pKa = 8.59VKK139 pKa = 10.69KK140 pKa = 10.82VEE142 pKa = 4.21DD143 pKa = 3.63KK144 pKa = 11.06RR145 pKa = 11.84AGTQTPNEE153 pKa = 4.1VALCQIFTMQQGIMAEE169 pKa = 4.18EE170 pKa = 4.24VKK172 pKa = 10.32EE173 pKa = 4.08ARR175 pKa = 11.84ASTQQEE181 pKa = 3.64IDD183 pKa = 3.19EE184 pKa = 4.33LTRR187 pKa = 11.84LLRR190 pKa = 11.84LKK192 pKa = 10.34RR193 pKa = 11.84AEE195 pKa = 4.0QVKK198 pKa = 10.22VLAEE202 pKa = 4.01IKK204 pKa = 10.48DD205 pKa = 3.61KK206 pKa = 11.31YY207 pKa = 10.69FPANIWEE214 pKa = 4.41EE215 pKa = 3.79PAEE218 pKa = 4.08AEE220 pKa = 4.08RR221 pKa = 11.84NARR224 pKa = 11.84CWEE227 pKa = 4.25VYY229 pKa = 10.07SAALVTAGRR238 pKa = 11.84PAPIKK243 pKa = 10.02TEE245 pKa = 3.85AAFKK249 pKa = 10.6LAIDD253 pKa = 4.29AYY255 pKa = 9.86KK256 pKa = 11.02NFVDD260 pKa = 4.13TEE262 pKa = 4.18FKK264 pKa = 10.52TRR266 pKa = 11.84FIRR269 pKa = 11.84SDD271 pKa = 3.14EE272 pKa = 4.18HH273 pKa = 7.42QVALRR278 pKa = 11.84KK279 pKa = 9.17FADD282 pKa = 3.64EE283 pKa = 5.12RR284 pKa = 11.84IHH286 pKa = 6.66FLDD289 pKa = 6.09GIGEE293 pKa = 4.17PKK295 pKa = 10.29KK296 pKa = 10.81AGTFRR301 pKa = 11.84SLLAVLGVEE310 pKa = 4.99PSAQTTTHH318 pKa = 5.1TEE320 pKa = 3.69EE321 pKa = 5.61RR322 pKa = 11.84NDD324 pKa = 3.55EE325 pKa = 4.29DD326 pKa = 5.44DD327 pKa = 4.46SGGEE331 pKa = 3.8SDD333 pKa = 3.7VARR336 pKa = 11.84GEE338 pKa = 4.16EE339 pKa = 4.32PEE341 pKa = 4.34PEE343 pKa = 4.38AGGGTSRR350 pKa = 11.84PGASGRR356 pKa = 11.84GAGRR360 pKa = 11.84GKK362 pKa = 10.52GKK364 pKa = 10.25RR365 pKa = 11.84KK366 pKa = 9.41VVPEE370 pKa = 3.84RR371 pKa = 11.84RR372 pKa = 11.84SLRR375 pKa = 11.84SSKK378 pKa = 9.29TGRR381 pKa = 11.84VTDD384 pKa = 3.53VSVQGAQGDD393 pKa = 4.3GGASRR398 pKa = 11.84PKK400 pKa = 9.9HH401 pKa = 5.49RR402 pKa = 11.84RR403 pKa = 11.84RR404 pKa = 3.44
Molecular weight: 44.22 kDa
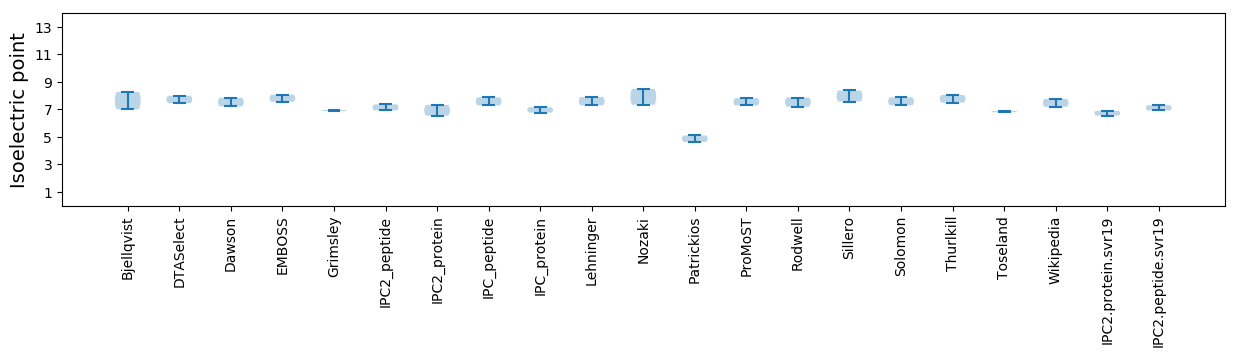
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E1B261|E1B261_9VIRU Putative coat protein OS=Rhododendron virus A OX=878260 PE=4 SV=1
MM1 pKa = 7.78EE2 pKa = 5.6DD3 pKa = 3.06RR4 pKa = 11.84RR5 pKa = 11.84AEE7 pKa = 4.13DD8 pKa = 4.46RR9 pKa = 11.84LDD11 pKa = 4.38DD12 pKa = 4.86DD13 pKa = 5.18LPAGGGAPPPLRR25 pKa = 11.84GLPPLPRR32 pKa = 11.84AEE34 pKa = 4.14PTDD37 pKa = 4.25AEE39 pKa = 4.13LQAAIDD45 pKa = 3.66EE46 pKa = 4.61AVAVVYY52 pKa = 9.38EE53 pKa = 4.19AGMPAGRR60 pKa = 11.84FTVQRR65 pKa = 11.84VHH67 pKa = 7.07EE68 pKa = 4.6IGLTVEE74 pKa = 4.45VFVKK78 pKa = 9.63QARR81 pKa = 11.84AVFGGKK87 pKa = 9.29NVDD90 pKa = 3.42QADD93 pKa = 4.34LIFTTGIKK101 pKa = 9.67MGVCGSLRR109 pKa = 11.84TMEE112 pKa = 4.42PAGFWEE118 pKa = 4.71VIRR121 pKa = 11.84WARR124 pKa = 11.84SNVGRR129 pKa = 11.84QALEE133 pKa = 3.77TGQKK137 pKa = 8.59VKK139 pKa = 10.69KK140 pKa = 10.82VEE142 pKa = 4.21DD143 pKa = 3.63KK144 pKa = 11.06RR145 pKa = 11.84AGTQTPNEE153 pKa = 4.1VALCQIFTMQQGIMAEE169 pKa = 4.18EE170 pKa = 4.24VKK172 pKa = 10.32EE173 pKa = 4.08ARR175 pKa = 11.84ASTQQEE181 pKa = 3.64IDD183 pKa = 3.19EE184 pKa = 4.33LTRR187 pKa = 11.84LLRR190 pKa = 11.84LKK192 pKa = 10.34RR193 pKa = 11.84AEE195 pKa = 4.0QVKK198 pKa = 10.22VLAEE202 pKa = 4.01IKK204 pKa = 10.48DD205 pKa = 3.61KK206 pKa = 11.31YY207 pKa = 10.69FPANIWEE214 pKa = 4.41EE215 pKa = 3.79PAEE218 pKa = 4.08AEE220 pKa = 4.08RR221 pKa = 11.84NARR224 pKa = 11.84CWEE227 pKa = 4.25VYY229 pKa = 10.07SAALVTAGRR238 pKa = 11.84PAPIKK243 pKa = 10.02TEE245 pKa = 3.85AAFKK249 pKa = 10.6LAIDD253 pKa = 4.29AYY255 pKa = 9.86KK256 pKa = 11.02NFVDD260 pKa = 4.13TEE262 pKa = 4.18FKK264 pKa = 10.52TRR266 pKa = 11.84FIRR269 pKa = 11.84SDD271 pKa = 3.14EE272 pKa = 4.18HH273 pKa = 7.42QVALRR278 pKa = 11.84KK279 pKa = 9.17FADD282 pKa = 3.64EE283 pKa = 5.12RR284 pKa = 11.84IHH286 pKa = 6.66FLDD289 pKa = 6.09GIGEE293 pKa = 4.17PKK295 pKa = 10.29KK296 pKa = 10.81AGTFRR301 pKa = 11.84SLLAVLGVEE310 pKa = 4.99PSAQTTTHH318 pKa = 5.1TEE320 pKa = 3.69EE321 pKa = 5.61RR322 pKa = 11.84NDD324 pKa = 3.55EE325 pKa = 4.29DD326 pKa = 5.44DD327 pKa = 4.46SGGEE331 pKa = 3.8SDD333 pKa = 3.7VARR336 pKa = 11.84GEE338 pKa = 4.16EE339 pKa = 4.32PEE341 pKa = 4.34PEE343 pKa = 4.38AGGGTSRR350 pKa = 11.84PGASGRR356 pKa = 11.84GAGRR360 pKa = 11.84GKK362 pKa = 10.95GNARR366 pKa = 11.84LSRR369 pKa = 11.84NVEE372 pKa = 3.43VSAQVRR378 pKa = 11.84LGEE381 pKa = 4.01LQTYY385 pKa = 8.15RR386 pKa = 11.84FRR388 pKa = 11.84VHH390 pKa = 7.71RR391 pKa = 11.84EE392 pKa = 3.76MVEE395 pKa = 3.62HH396 pKa = 6.74RR397 pKa = 11.84VRR399 pKa = 11.84SIAVARR405 pKa = 11.84SRR407 pKa = 11.84FEE409 pKa = 3.66AGIRR413 pKa = 11.84RR414 pKa = 11.84IIGGGEE420 pKa = 3.64MRR422 pKa = 11.84SWYY425 pKa = 9.02TDD427 pKa = 2.35SRR429 pKa = 11.84MYY431 pKa = 10.69RR432 pKa = 11.84GGGNSNDD439 pKa = 3.26AMKK442 pKa = 10.46LLSAASVVTPGKK454 pKa = 10.44FLDD457 pKa = 3.1QCYY460 pKa = 9.29RR461 pKa = 11.84VSSARR466 pKa = 11.84KK467 pKa = 9.09ALRR470 pKa = 11.84LPSGLRR476 pKa = 11.84VPDD479 pKa = 3.96SLAEE483 pKa = 4.24CQCKK487 pKa = 8.36TFNDD491 pKa = 3.69QASAGPFLRR500 pKa = 11.84AFGIKK505 pKa = 9.53KK506 pKa = 10.18KK507 pKa = 10.4YY508 pKa = 9.44GLRR511 pKa = 11.84RR512 pKa = 11.84MLEE515 pKa = 3.73EE516 pKa = 4.28FMWGIYY522 pKa = 9.83DD523 pKa = 3.73RR524 pKa = 11.84YY525 pKa = 10.54GDD527 pKa = 3.54GRR529 pKa = 11.84GDD531 pKa = 3.49EE532 pKa = 4.19RR533 pKa = 11.84SLPFITGRR541 pKa = 11.84IGFRR545 pKa = 11.84TKK547 pKa = 10.67LLEE550 pKa = 4.26EE551 pKa = 4.75GEE553 pKa = 4.39ALRR556 pKa = 11.84KK557 pKa = 8.94IRR559 pKa = 11.84NGEE562 pKa = 3.83PLGRR566 pKa = 11.84AVMMLDD572 pKa = 3.21ALEE575 pKa = 4.77QISSSPLYY583 pKa = 10.49NILSGYY589 pKa = 9.64CGRR592 pKa = 11.84NRR594 pKa = 11.84NKK596 pKa = 9.97GWCEE600 pKa = 3.55FRR602 pKa = 11.84NTTVRR607 pKa = 11.84ASSDD611 pKa = 2.72WGLLYY616 pKa = 10.9SEE618 pKa = 4.79VEE620 pKa = 3.84RR621 pKa = 11.84AKK623 pKa = 10.95CIVEE627 pKa = 4.21LDD629 pKa = 3.0WSKK632 pKa = 10.98FDD634 pKa = 3.92RR635 pKa = 11.84EE636 pKa = 4.15RR637 pKa = 11.84PSRR640 pKa = 11.84DD641 pKa = 2.65ILFVIKK647 pKa = 10.31VIISCFEE654 pKa = 3.81PRR656 pKa = 11.84NGRR659 pKa = 11.84EE660 pKa = 3.61RR661 pKa = 11.84RR662 pKa = 11.84LLEE665 pKa = 4.13AYY667 pKa = 10.01EE668 pKa = 4.08IMLEE672 pKa = 3.9RR673 pKa = 11.84ALVHH677 pKa = 7.07RR678 pKa = 11.84LLLTDD683 pKa = 3.78AGGVLEE689 pKa = 4.47MEE691 pKa = 4.59GMVPSGSLWTGWLDD705 pKa = 3.23TAMNILYY712 pKa = 9.79IRR714 pKa = 11.84AACLGAGIASDD725 pKa = 4.99LYY727 pKa = 11.14SSRR730 pKa = 11.84CAGDD734 pKa = 4.48DD735 pKa = 3.47NLTLFWEE742 pKa = 4.63DD743 pKa = 3.25QSDD746 pKa = 4.52GILGLVKK753 pKa = 10.19QHH755 pKa = 6.07LNNWFRR761 pKa = 11.84AGIKK765 pKa = 10.55DD766 pKa = 3.61EE767 pKa = 5.05DD768 pKa = 4.61FIICRR773 pKa = 11.84GPYY776 pKa = 9.02YY777 pKa = 10.15VEE779 pKa = 5.41RR780 pKa = 11.84YY781 pKa = 8.48QATFPIGTDD790 pKa = 3.26LSEE793 pKa = 4.36GTSRR797 pKa = 11.84LMDD800 pKa = 3.42SAIWVRR806 pKa = 11.84LEE808 pKa = 3.86GGPIIDD814 pKa = 3.84ANRR817 pKa = 11.84GLSHH821 pKa = 6.37RR822 pKa = 11.84WEE824 pKa = 4.12YY825 pKa = 11.01RR826 pKa = 11.84FHH828 pKa = 7.2GKK830 pKa = 9.25PKK832 pKa = 10.31FLSCYY837 pKa = 8.02WLPDD841 pKa = 3.23GRR843 pKa = 11.84PIRR846 pKa = 11.84PAHH849 pKa = 6.66DD850 pKa = 3.97CLEE853 pKa = 4.17KK854 pKa = 11.01LLWPEE859 pKa = 5.03GIHH862 pKa = 7.16GDD864 pKa = 3.83LEE866 pKa = 4.49TYY868 pKa = 8.56EE869 pKa = 4.24AAVISMIVDD878 pKa = 3.26NPFNHH883 pKa = 6.73HH884 pKa = 6.14NVNHH888 pKa = 5.58MLVRR892 pKa = 11.84YY893 pKa = 9.31VLIQQIKK900 pKa = 7.99RR901 pKa = 11.84QVVSPMTAEE910 pKa = 4.87DD911 pKa = 3.38IVYY914 pKa = 9.97LCKK917 pKa = 10.52FRR919 pKa = 11.84DD920 pKa = 3.69RR921 pKa = 11.84EE922 pKa = 4.45GGSVPYY928 pKa = 10.42PMIGPWRR935 pKa = 11.84RR936 pKa = 11.84GQKK939 pKa = 8.65QGRR942 pKa = 11.84MEE944 pKa = 5.31DD945 pKa = 3.81YY946 pKa = 10.69PEE948 pKa = 3.94VGRR951 pKa = 11.84DD952 pKa = 3.03IKK954 pKa = 10.92NLKK957 pKa = 10.06GFVAGISTLYY967 pKa = 9.94ARR969 pKa = 11.84KK970 pKa = 10.09GGGGIDD976 pKa = 2.85AWRR979 pKa = 11.84FMDD982 pKa = 5.64IIRR985 pKa = 11.84GDD987 pKa = 3.45ADD989 pKa = 3.81LGQGQFGNEE998 pKa = 3.74VLEE1001 pKa = 4.09WARR1004 pKa = 11.84WLGRR1008 pKa = 11.84HH1009 pKa = 5.47EE1010 pKa = 4.1VTKK1013 pKa = 10.05FLRR1016 pKa = 11.84PIKK1019 pKa = 10.24RR1020 pKa = 11.84MRR1022 pKa = 11.84AEE1024 pKa = 3.95HH1025 pKa = 6.55GEE1027 pKa = 4.13VIIEE1031 pKa = 4.19GEE1033 pKa = 4.17GRR1035 pKa = 11.84RR1036 pKa = 11.84QAEE1039 pKa = 3.75AALRR1043 pKa = 11.84ALRR1046 pKa = 11.84EE1047 pKa = 3.86ILNEE1051 pKa = 3.96GTHH1054 pKa = 6.65RR1055 pKa = 11.84SCLDD1059 pKa = 3.13FSFFLSDD1066 pKa = 3.84RR1067 pKa = 11.84LRR1069 pKa = 11.84ASCEE1073 pKa = 3.71DD1074 pKa = 3.41PRR1076 pKa = 11.84RR1077 pKa = 3.67
MM1 pKa = 7.78EE2 pKa = 5.6DD3 pKa = 3.06RR4 pKa = 11.84RR5 pKa = 11.84AEE7 pKa = 4.13DD8 pKa = 4.46RR9 pKa = 11.84LDD11 pKa = 4.38DD12 pKa = 4.86DD13 pKa = 5.18LPAGGGAPPPLRR25 pKa = 11.84GLPPLPRR32 pKa = 11.84AEE34 pKa = 4.14PTDD37 pKa = 4.25AEE39 pKa = 4.13LQAAIDD45 pKa = 3.66EE46 pKa = 4.61AVAVVYY52 pKa = 9.38EE53 pKa = 4.19AGMPAGRR60 pKa = 11.84FTVQRR65 pKa = 11.84VHH67 pKa = 7.07EE68 pKa = 4.6IGLTVEE74 pKa = 4.45VFVKK78 pKa = 9.63QARR81 pKa = 11.84AVFGGKK87 pKa = 9.29NVDD90 pKa = 3.42QADD93 pKa = 4.34LIFTTGIKK101 pKa = 9.67MGVCGSLRR109 pKa = 11.84TMEE112 pKa = 4.42PAGFWEE118 pKa = 4.71VIRR121 pKa = 11.84WARR124 pKa = 11.84SNVGRR129 pKa = 11.84QALEE133 pKa = 3.77TGQKK137 pKa = 8.59VKK139 pKa = 10.69KK140 pKa = 10.82VEE142 pKa = 4.21DD143 pKa = 3.63KK144 pKa = 11.06RR145 pKa = 11.84AGTQTPNEE153 pKa = 4.1VALCQIFTMQQGIMAEE169 pKa = 4.18EE170 pKa = 4.24VKK172 pKa = 10.32EE173 pKa = 4.08ARR175 pKa = 11.84ASTQQEE181 pKa = 3.64IDD183 pKa = 3.19EE184 pKa = 4.33LTRR187 pKa = 11.84LLRR190 pKa = 11.84LKK192 pKa = 10.34RR193 pKa = 11.84AEE195 pKa = 4.0QVKK198 pKa = 10.22VLAEE202 pKa = 4.01IKK204 pKa = 10.48DD205 pKa = 3.61KK206 pKa = 11.31YY207 pKa = 10.69FPANIWEE214 pKa = 4.41EE215 pKa = 3.79PAEE218 pKa = 4.08AEE220 pKa = 4.08RR221 pKa = 11.84NARR224 pKa = 11.84CWEE227 pKa = 4.25VYY229 pKa = 10.07SAALVTAGRR238 pKa = 11.84PAPIKK243 pKa = 10.02TEE245 pKa = 3.85AAFKK249 pKa = 10.6LAIDD253 pKa = 4.29AYY255 pKa = 9.86KK256 pKa = 11.02NFVDD260 pKa = 4.13TEE262 pKa = 4.18FKK264 pKa = 10.52TRR266 pKa = 11.84FIRR269 pKa = 11.84SDD271 pKa = 3.14EE272 pKa = 4.18HH273 pKa = 7.42QVALRR278 pKa = 11.84KK279 pKa = 9.17FADD282 pKa = 3.64EE283 pKa = 5.12RR284 pKa = 11.84IHH286 pKa = 6.66FLDD289 pKa = 6.09GIGEE293 pKa = 4.17PKK295 pKa = 10.29KK296 pKa = 10.81AGTFRR301 pKa = 11.84SLLAVLGVEE310 pKa = 4.99PSAQTTTHH318 pKa = 5.1TEE320 pKa = 3.69EE321 pKa = 5.61RR322 pKa = 11.84NDD324 pKa = 3.55EE325 pKa = 4.29DD326 pKa = 5.44DD327 pKa = 4.46SGGEE331 pKa = 3.8SDD333 pKa = 3.7VARR336 pKa = 11.84GEE338 pKa = 4.16EE339 pKa = 4.32PEE341 pKa = 4.34PEE343 pKa = 4.38AGGGTSRR350 pKa = 11.84PGASGRR356 pKa = 11.84GAGRR360 pKa = 11.84GKK362 pKa = 10.95GNARR366 pKa = 11.84LSRR369 pKa = 11.84NVEE372 pKa = 3.43VSAQVRR378 pKa = 11.84LGEE381 pKa = 4.01LQTYY385 pKa = 8.15RR386 pKa = 11.84FRR388 pKa = 11.84VHH390 pKa = 7.71RR391 pKa = 11.84EE392 pKa = 3.76MVEE395 pKa = 3.62HH396 pKa = 6.74RR397 pKa = 11.84VRR399 pKa = 11.84SIAVARR405 pKa = 11.84SRR407 pKa = 11.84FEE409 pKa = 3.66AGIRR413 pKa = 11.84RR414 pKa = 11.84IIGGGEE420 pKa = 3.64MRR422 pKa = 11.84SWYY425 pKa = 9.02TDD427 pKa = 2.35SRR429 pKa = 11.84MYY431 pKa = 10.69RR432 pKa = 11.84GGGNSNDD439 pKa = 3.26AMKK442 pKa = 10.46LLSAASVVTPGKK454 pKa = 10.44FLDD457 pKa = 3.1QCYY460 pKa = 9.29RR461 pKa = 11.84VSSARR466 pKa = 11.84KK467 pKa = 9.09ALRR470 pKa = 11.84LPSGLRR476 pKa = 11.84VPDD479 pKa = 3.96SLAEE483 pKa = 4.24CQCKK487 pKa = 8.36TFNDD491 pKa = 3.69QASAGPFLRR500 pKa = 11.84AFGIKK505 pKa = 9.53KK506 pKa = 10.18KK507 pKa = 10.4YY508 pKa = 9.44GLRR511 pKa = 11.84RR512 pKa = 11.84MLEE515 pKa = 3.73EE516 pKa = 4.28FMWGIYY522 pKa = 9.83DD523 pKa = 3.73RR524 pKa = 11.84YY525 pKa = 10.54GDD527 pKa = 3.54GRR529 pKa = 11.84GDD531 pKa = 3.49EE532 pKa = 4.19RR533 pKa = 11.84SLPFITGRR541 pKa = 11.84IGFRR545 pKa = 11.84TKK547 pKa = 10.67LLEE550 pKa = 4.26EE551 pKa = 4.75GEE553 pKa = 4.39ALRR556 pKa = 11.84KK557 pKa = 8.94IRR559 pKa = 11.84NGEE562 pKa = 3.83PLGRR566 pKa = 11.84AVMMLDD572 pKa = 3.21ALEE575 pKa = 4.77QISSSPLYY583 pKa = 10.49NILSGYY589 pKa = 9.64CGRR592 pKa = 11.84NRR594 pKa = 11.84NKK596 pKa = 9.97GWCEE600 pKa = 3.55FRR602 pKa = 11.84NTTVRR607 pKa = 11.84ASSDD611 pKa = 2.72WGLLYY616 pKa = 10.9SEE618 pKa = 4.79VEE620 pKa = 3.84RR621 pKa = 11.84AKK623 pKa = 10.95CIVEE627 pKa = 4.21LDD629 pKa = 3.0WSKK632 pKa = 10.98FDD634 pKa = 3.92RR635 pKa = 11.84EE636 pKa = 4.15RR637 pKa = 11.84PSRR640 pKa = 11.84DD641 pKa = 2.65ILFVIKK647 pKa = 10.31VIISCFEE654 pKa = 3.81PRR656 pKa = 11.84NGRR659 pKa = 11.84EE660 pKa = 3.61RR661 pKa = 11.84RR662 pKa = 11.84LLEE665 pKa = 4.13AYY667 pKa = 10.01EE668 pKa = 4.08IMLEE672 pKa = 3.9RR673 pKa = 11.84ALVHH677 pKa = 7.07RR678 pKa = 11.84LLLTDD683 pKa = 3.78AGGVLEE689 pKa = 4.47MEE691 pKa = 4.59GMVPSGSLWTGWLDD705 pKa = 3.23TAMNILYY712 pKa = 9.79IRR714 pKa = 11.84AACLGAGIASDD725 pKa = 4.99LYY727 pKa = 11.14SSRR730 pKa = 11.84CAGDD734 pKa = 4.48DD735 pKa = 3.47NLTLFWEE742 pKa = 4.63DD743 pKa = 3.25QSDD746 pKa = 4.52GILGLVKK753 pKa = 10.19QHH755 pKa = 6.07LNNWFRR761 pKa = 11.84AGIKK765 pKa = 10.55DD766 pKa = 3.61EE767 pKa = 5.05DD768 pKa = 4.61FIICRR773 pKa = 11.84GPYY776 pKa = 9.02YY777 pKa = 10.15VEE779 pKa = 5.41RR780 pKa = 11.84YY781 pKa = 8.48QATFPIGTDD790 pKa = 3.26LSEE793 pKa = 4.36GTSRR797 pKa = 11.84LMDD800 pKa = 3.42SAIWVRR806 pKa = 11.84LEE808 pKa = 3.86GGPIIDD814 pKa = 3.84ANRR817 pKa = 11.84GLSHH821 pKa = 6.37RR822 pKa = 11.84WEE824 pKa = 4.12YY825 pKa = 11.01RR826 pKa = 11.84FHH828 pKa = 7.2GKK830 pKa = 9.25PKK832 pKa = 10.31FLSCYY837 pKa = 8.02WLPDD841 pKa = 3.23GRR843 pKa = 11.84PIRR846 pKa = 11.84PAHH849 pKa = 6.66DD850 pKa = 3.97CLEE853 pKa = 4.17KK854 pKa = 11.01LLWPEE859 pKa = 5.03GIHH862 pKa = 7.16GDD864 pKa = 3.83LEE866 pKa = 4.49TYY868 pKa = 8.56EE869 pKa = 4.24AAVISMIVDD878 pKa = 3.26NPFNHH883 pKa = 6.73HH884 pKa = 6.14NVNHH888 pKa = 5.58MLVRR892 pKa = 11.84YY893 pKa = 9.31VLIQQIKK900 pKa = 7.99RR901 pKa = 11.84QVVSPMTAEE910 pKa = 4.87DD911 pKa = 3.38IVYY914 pKa = 9.97LCKK917 pKa = 10.52FRR919 pKa = 11.84DD920 pKa = 3.69RR921 pKa = 11.84EE922 pKa = 4.45GGSVPYY928 pKa = 10.42PMIGPWRR935 pKa = 11.84RR936 pKa = 11.84GQKK939 pKa = 8.65QGRR942 pKa = 11.84MEE944 pKa = 5.31DD945 pKa = 3.81YY946 pKa = 10.69PEE948 pKa = 3.94VGRR951 pKa = 11.84DD952 pKa = 3.03IKK954 pKa = 10.92NLKK957 pKa = 10.06GFVAGISTLYY967 pKa = 9.94ARR969 pKa = 11.84KK970 pKa = 10.09GGGGIDD976 pKa = 2.85AWRR979 pKa = 11.84FMDD982 pKa = 5.64IIRR985 pKa = 11.84GDD987 pKa = 3.45ADD989 pKa = 3.81LGQGQFGNEE998 pKa = 3.74VLEE1001 pKa = 4.09WARR1004 pKa = 11.84WLGRR1008 pKa = 11.84HH1009 pKa = 5.47EE1010 pKa = 4.1VTKK1013 pKa = 10.05FLRR1016 pKa = 11.84PIKK1019 pKa = 10.24RR1020 pKa = 11.84MRR1022 pKa = 11.84AEE1024 pKa = 3.95HH1025 pKa = 6.55GEE1027 pKa = 4.13VIIEE1031 pKa = 4.19GEE1033 pKa = 4.17GRR1035 pKa = 11.84RR1036 pKa = 11.84QAEE1039 pKa = 3.75AALRR1043 pKa = 11.84ALRR1046 pKa = 11.84EE1047 pKa = 3.86ILNEE1051 pKa = 3.96GTHH1054 pKa = 6.65RR1055 pKa = 11.84SCLDD1059 pKa = 3.13FSFFLSDD1066 pKa = 3.84RR1067 pKa = 11.84LRR1069 pKa = 11.84ASCEE1073 pKa = 3.71DD1074 pKa = 3.41PRR1076 pKa = 11.84RR1077 pKa = 3.67
Molecular weight: 121.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1481 |
404 |
1077 |
740.5 |
82.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.386 ± 1.094 | 1.418 ± 0.329 |
5.672 ± 0.01 | 8.845 ± 0.514 |
3.781 ± 0.154 | 9.588 ± 0.032 |
1.553 ± 0.153 | 5.064 ± 0.657 |
4.727 ± 0.591 | 7.833 ± 0.92 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.161 ± 0.329 | 2.498 ± 0.372 |
4.591 ± 0.415 | 3.309 ± 0.438 |
10.128 ± 0.231 | 4.794 ± 0.406 |
4.456 ± 0.722 | 6.347 ± 0.645 |
1.688 ± 0.34 | 2.161 ± 0.569 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
