
Drosophila melanogaster sigma virus (isolate Drosophila/USA/AP30/2005) (DMelSV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Sigmavirus; Drosophila melanogaster sigmavirus
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
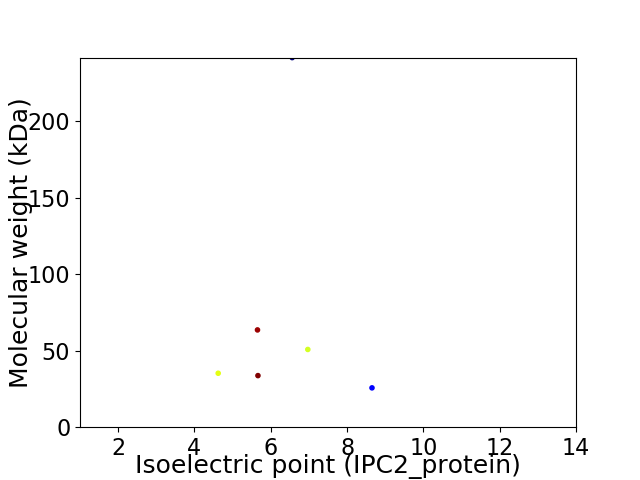
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|A7WNB3|GLYCO_DMSVA Glycoprotein OS=Drosophila melanogaster sigma virus (isolate Drosophila/USA/AP30/2005) OX=666363 GN=G PE=3 SV=1
MM1 pKa = 7.54NLTQEE6 pKa = 4.33DD7 pKa = 4.31EE8 pKa = 4.08QRR10 pKa = 11.84LALLRR15 pKa = 11.84EE16 pKa = 4.12RR17 pKa = 11.84AAAWDD22 pKa = 3.63HH23 pKa = 6.46KK24 pKa = 10.8NALGEE29 pKa = 4.23FEE31 pKa = 5.81SNAEE35 pKa = 3.64LDD37 pKa = 3.88VYY39 pKa = 11.37NEE41 pKa = 4.62GIAYY45 pKa = 7.87PPSIDD50 pKa = 3.28EE51 pKa = 4.33MEE53 pKa = 4.33VGEE56 pKa = 5.36GRR58 pKa = 11.84PTLDD62 pKa = 4.73KK63 pKa = 10.76ILEE66 pKa = 4.1SSIGSDD72 pKa = 3.35SLPNLEE78 pKa = 5.66DD79 pKa = 4.12GPPDD83 pKa = 3.29QEE85 pKa = 4.05AAYY88 pKa = 10.18HH89 pKa = 5.52EE90 pKa = 4.71GEE92 pKa = 4.52MYY94 pKa = 9.47STMMTSYY101 pKa = 10.32MDD103 pKa = 3.51QDD105 pKa = 4.17NEE107 pKa = 4.53NMDD110 pKa = 4.49LSAGPTCSTPQPITGIHH127 pKa = 6.11HH128 pKa = 6.96KK129 pKa = 8.77EE130 pKa = 4.18TINGRR135 pKa = 11.84VTQMVYY141 pKa = 10.01IGEE144 pKa = 4.29TSDD147 pKa = 3.03MALINRR153 pKa = 11.84LQDD156 pKa = 3.57EE157 pKa = 4.43FDD159 pKa = 3.56VLYY162 pKa = 9.15NTYY165 pKa = 10.6PNRR168 pKa = 11.84NKK170 pKa = 10.18QGEE173 pKa = 4.4IVHH176 pKa = 6.68LIVMWSTPLKK186 pKa = 10.1PSTRR190 pKa = 11.84YY191 pKa = 9.89SLSPAQPPGKK201 pKa = 9.49KK202 pKa = 9.58RR203 pKa = 11.84KK204 pKa = 9.05IVQHH208 pKa = 5.96SSNPEE213 pKa = 3.65PEE215 pKa = 4.06IVFVPEE221 pKa = 4.2PTSSEE226 pKa = 3.6QSLPEE231 pKa = 4.01RR232 pKa = 11.84QIKK235 pKa = 10.25AYY237 pKa = 8.41LQRR240 pKa = 11.84GIEE243 pKa = 4.16LKK245 pKa = 10.67GKK247 pKa = 9.54SAEE250 pKa = 4.11VPNFKK255 pKa = 8.75ITNEE259 pKa = 4.01TLGFSDD265 pKa = 5.4ANLKK269 pKa = 10.15LLYY272 pKa = 9.26PDD274 pKa = 3.44TSLYY278 pKa = 9.8PDD280 pKa = 3.49NPVFILEE287 pKa = 4.57DD288 pKa = 3.48VFASTRR294 pKa = 11.84QLNKK298 pKa = 10.15IKK300 pKa = 10.79VNYY303 pKa = 9.29QLYY306 pKa = 10.64DD307 pKa = 3.47PVLPSII313 pKa = 4.68
MM1 pKa = 7.54NLTQEE6 pKa = 4.33DD7 pKa = 4.31EE8 pKa = 4.08QRR10 pKa = 11.84LALLRR15 pKa = 11.84EE16 pKa = 4.12RR17 pKa = 11.84AAAWDD22 pKa = 3.63HH23 pKa = 6.46KK24 pKa = 10.8NALGEE29 pKa = 4.23FEE31 pKa = 5.81SNAEE35 pKa = 3.64LDD37 pKa = 3.88VYY39 pKa = 11.37NEE41 pKa = 4.62GIAYY45 pKa = 7.87PPSIDD50 pKa = 3.28EE51 pKa = 4.33MEE53 pKa = 4.33VGEE56 pKa = 5.36GRR58 pKa = 11.84PTLDD62 pKa = 4.73KK63 pKa = 10.76ILEE66 pKa = 4.1SSIGSDD72 pKa = 3.35SLPNLEE78 pKa = 5.66DD79 pKa = 4.12GPPDD83 pKa = 3.29QEE85 pKa = 4.05AAYY88 pKa = 10.18HH89 pKa = 5.52EE90 pKa = 4.71GEE92 pKa = 4.52MYY94 pKa = 9.47STMMTSYY101 pKa = 10.32MDD103 pKa = 3.51QDD105 pKa = 4.17NEE107 pKa = 4.53NMDD110 pKa = 4.49LSAGPTCSTPQPITGIHH127 pKa = 6.11HH128 pKa = 6.96KK129 pKa = 8.77EE130 pKa = 4.18TINGRR135 pKa = 11.84VTQMVYY141 pKa = 10.01IGEE144 pKa = 4.29TSDD147 pKa = 3.03MALINRR153 pKa = 11.84LQDD156 pKa = 3.57EE157 pKa = 4.43FDD159 pKa = 3.56VLYY162 pKa = 9.15NTYY165 pKa = 10.6PNRR168 pKa = 11.84NKK170 pKa = 10.18QGEE173 pKa = 4.4IVHH176 pKa = 6.68LIVMWSTPLKK186 pKa = 10.1PSTRR190 pKa = 11.84YY191 pKa = 9.89SLSPAQPPGKK201 pKa = 9.49KK202 pKa = 9.58RR203 pKa = 11.84KK204 pKa = 9.05IVQHH208 pKa = 5.96SSNPEE213 pKa = 3.65PEE215 pKa = 4.06IVFVPEE221 pKa = 4.2PTSSEE226 pKa = 3.6QSLPEE231 pKa = 4.01RR232 pKa = 11.84QIKK235 pKa = 10.25AYY237 pKa = 8.41LQRR240 pKa = 11.84GIEE243 pKa = 4.16LKK245 pKa = 10.67GKK247 pKa = 9.54SAEE250 pKa = 4.11VPNFKK255 pKa = 8.75ITNEE259 pKa = 4.01TLGFSDD265 pKa = 5.4ANLKK269 pKa = 10.15LLYY272 pKa = 9.26PDD274 pKa = 3.44TSLYY278 pKa = 9.8PDD280 pKa = 3.49NPVFILEE287 pKa = 4.57DD288 pKa = 3.48VFASTRR294 pKa = 11.84QLNKK298 pKa = 10.15IKK300 pKa = 10.79VNYY303 pKa = 9.29QLYY306 pKa = 10.64DD307 pKa = 3.47PVLPSII313 pKa = 4.68
Molecular weight: 35.25 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|A7WNB2|PHOSP_DMSVA Phosphoprotein OS=Drosophila melanogaster sigma virus (isolate Drosophila/USA/AP30/2005) OX=666363 GN=P PE=3 SV=2
MM1 pKa = 7.43NKK3 pKa = 9.12MNQLVRR9 pKa = 11.84FVKK12 pKa = 10.03DD13 pKa = 3.36TVAVRR18 pKa = 11.84KK19 pKa = 8.24PQSEE23 pKa = 4.34DD24 pKa = 2.85KK25 pKa = 10.77SYY27 pKa = 11.55LPIPSTIGGHH37 pKa = 5.16EE38 pKa = 4.31VNSPFAEE45 pKa = 4.11PTAPSLGIIQPKK57 pKa = 9.46CKK59 pKa = 10.12RR60 pKa = 11.84ADD62 pKa = 3.41WLIKK66 pKa = 10.12SHH68 pKa = 6.41LTITTNYY75 pKa = 9.13EE76 pKa = 3.56IKK78 pKa = 10.27EE79 pKa = 4.06WEE81 pKa = 4.01TWDD84 pKa = 4.09RR85 pKa = 11.84AISDD89 pKa = 3.89ILDD92 pKa = 4.21LYY94 pKa = 11.07DD95 pKa = 4.0GNPVFKK101 pKa = 10.42PILLFVYY108 pKa = 10.2YY109 pKa = 10.35VLAYY113 pKa = 9.41NARR116 pKa = 11.84KK117 pKa = 9.66IPGPSNGVRR126 pKa = 11.84YY127 pKa = 8.85GAYY130 pKa = 9.82FDD132 pKa = 4.64EE133 pKa = 5.29LTTVWHH139 pKa = 7.39AIPEE143 pKa = 4.24LMNQEE148 pKa = 3.08IDD150 pKa = 3.12YY151 pKa = 10.44SYY153 pKa = 10.98NHH155 pKa = 6.79RR156 pKa = 11.84VLHH159 pKa = 6.44RR160 pKa = 11.84KK161 pKa = 7.27IQYY164 pKa = 9.5VISFKK169 pKa = 10.67IQMSSTKK176 pKa = 10.13RR177 pKa = 11.84RR178 pKa = 11.84TSPIEE183 pKa = 3.66SFIEE187 pKa = 4.23VTSEE191 pKa = 4.07GLKK194 pKa = 8.66HH195 pKa = 5.87TPQFTTILDD204 pKa = 3.58RR205 pKa = 11.84ARR207 pKa = 11.84FVYY210 pKa = 10.56SLTGGRR216 pKa = 11.84YY217 pKa = 9.15VIHH220 pKa = 7.08PFF222 pKa = 3.4
MM1 pKa = 7.43NKK3 pKa = 9.12MNQLVRR9 pKa = 11.84FVKK12 pKa = 10.03DD13 pKa = 3.36TVAVRR18 pKa = 11.84KK19 pKa = 8.24PQSEE23 pKa = 4.34DD24 pKa = 2.85KK25 pKa = 10.77SYY27 pKa = 11.55LPIPSTIGGHH37 pKa = 5.16EE38 pKa = 4.31VNSPFAEE45 pKa = 4.11PTAPSLGIIQPKK57 pKa = 9.46CKK59 pKa = 10.12RR60 pKa = 11.84ADD62 pKa = 3.41WLIKK66 pKa = 10.12SHH68 pKa = 6.41LTITTNYY75 pKa = 9.13EE76 pKa = 3.56IKK78 pKa = 10.27EE79 pKa = 4.06WEE81 pKa = 4.01TWDD84 pKa = 4.09RR85 pKa = 11.84AISDD89 pKa = 3.89ILDD92 pKa = 4.21LYY94 pKa = 11.07DD95 pKa = 4.0GNPVFKK101 pKa = 10.42PILLFVYY108 pKa = 10.2YY109 pKa = 10.35VLAYY113 pKa = 9.41NARR116 pKa = 11.84KK117 pKa = 9.66IPGPSNGVRR126 pKa = 11.84YY127 pKa = 8.85GAYY130 pKa = 9.82FDD132 pKa = 4.64EE133 pKa = 5.29LTTVWHH139 pKa = 7.39AIPEE143 pKa = 4.24LMNQEE148 pKa = 3.08IDD150 pKa = 3.12YY151 pKa = 10.44SYY153 pKa = 10.98NHH155 pKa = 6.79RR156 pKa = 11.84VLHH159 pKa = 6.44RR160 pKa = 11.84KK161 pKa = 7.27IQYY164 pKa = 9.5VISFKK169 pKa = 10.67IQMSSTKK176 pKa = 10.13RR177 pKa = 11.84RR178 pKa = 11.84TSPIEE183 pKa = 3.66SFIEE187 pKa = 4.23VTSEE191 pKa = 4.07GLKK194 pKa = 8.66HH195 pKa = 5.87TPQFTTILDD204 pKa = 3.58RR205 pKa = 11.84ARR207 pKa = 11.84FVYY210 pKa = 10.56SLTGGRR216 pKa = 11.84YY217 pKa = 9.15VIHH220 pKa = 7.08PFF222 pKa = 3.4
Molecular weight: 25.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3973 |
222 |
2129 |
662.2 |
75.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.531 ± 0.331 | 1.435 ± 0.246 |
5.462 ± 0.337 | 5.613 ± 0.57 |
4.027 ± 0.358 | 5.965 ± 0.446 |
2.794 ± 0.25 | 6.746 ± 0.3 |
4.757 ± 0.374 | 9.791 ± 0.584 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.794 ± 0.129 | 4.656 ± 0.228 |
5.412 ± 0.482 | 3.624 ± 0.142 |
5.437 ± 0.355 | 8.205 ± 0.456 |
6.771 ± 0.513 | 6.443 ± 0.481 |
1.611 ± 0.34 | 3.927 ± 0.295 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
