
Beauveria bassiana polymycovirus 1
Taxonomy: Viruses; Riboviria; Polymycoviridae; Polymycovirus
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
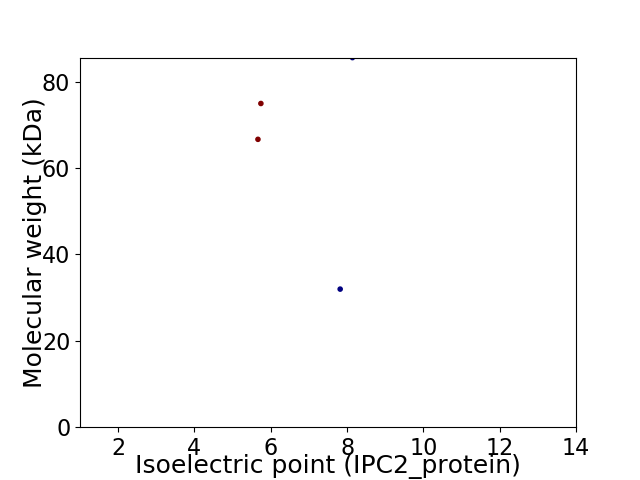
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1V1IEY9|A0A1V1IEY9_9VIRU RNA dependent RNA polymerase OS=Beauveria bassiana polymycovirus 1 OX=1740646 GN=RdRp PE=4 SV=1
MM1 pKa = 7.89ADD3 pKa = 3.79LTRR6 pKa = 11.84LSAIAVDD13 pKa = 4.05VGSPAPAVAAVLDD26 pKa = 4.18LFGVSDD32 pKa = 3.81PTGQCLSSAQVMSVVEE48 pKa = 4.73WIAGFPLQSAPGLMPAYY65 pKa = 9.55VFEE68 pKa = 4.37YY69 pKa = 10.21VKK71 pKa = 10.92SKK73 pKa = 10.86AGSLVASAGVCDD85 pKa = 4.07DD86 pKa = 2.96VAYY89 pKa = 10.26FLRR92 pKa = 11.84HH93 pKa = 5.74FEE95 pKa = 4.87APDD98 pKa = 3.1EE99 pKa = 4.9HH100 pKa = 6.07YY101 pKa = 9.28TKK103 pKa = 10.22SGRR106 pKa = 11.84SARR109 pKa = 11.84VAQVALNKK117 pKa = 9.41PLKK120 pKa = 10.61GDD122 pKa = 3.81GAHH125 pKa = 6.94LGAVEE130 pKa = 4.76LALLDD135 pKa = 3.56AVTRR139 pKa = 11.84VGYY142 pKa = 9.93VGPAGALASVARR154 pKa = 11.84LRR156 pKa = 11.84TMEE159 pKa = 4.11VLRR162 pKa = 11.84HH163 pKa = 4.85YY164 pKa = 10.95AVDD167 pKa = 3.42VRR169 pKa = 11.84QRR171 pKa = 11.84ACAQYY176 pKa = 10.67RR177 pKa = 11.84VTCQVCLPGRR187 pKa = 11.84DD188 pKa = 3.82SYY190 pKa = 11.84PLITPWCVSRR200 pKa = 11.84QSATICAGVILASVGGEE217 pKa = 3.64AATKK221 pKa = 9.61AVQRR225 pKa = 11.84RR226 pKa = 11.84AVTVASHH233 pKa = 7.06AARR236 pKa = 11.84IGLDD240 pKa = 3.1AVRR243 pKa = 11.84AIVAPACRR251 pKa = 11.84LVAEE255 pKa = 4.44YY256 pKa = 11.03SFDD259 pKa = 3.68PVRR262 pKa = 11.84LVLTDD267 pKa = 3.5RR268 pKa = 11.84YY269 pKa = 10.28GRR271 pKa = 11.84VRR273 pKa = 11.84EE274 pKa = 4.38RR275 pKa = 11.84ITSRR279 pKa = 11.84SPAATVAFAYY289 pKa = 7.65LTSRR293 pKa = 11.84GSADD297 pKa = 3.79PLLAMASEE305 pKa = 4.56RR306 pKa = 11.84VEE308 pKa = 4.08RR309 pKa = 11.84MGPVEE314 pKa = 5.63LLPQCVQDD322 pKa = 3.3YY323 pKa = 9.09WNQSEE328 pKa = 5.53DD329 pKa = 3.49IVGMLSLFCRR339 pKa = 11.84ALHH342 pKa = 5.17LTRR345 pKa = 11.84VEE347 pKa = 4.01RR348 pKa = 11.84EE349 pKa = 3.68EE350 pKa = 4.05EE351 pKa = 3.85EE352 pKa = 4.31RR353 pKa = 11.84GRR355 pKa = 11.84DD356 pKa = 3.77VNKK359 pKa = 10.41GPCLGYY365 pKa = 10.38LAVGRR370 pKa = 11.84KK371 pKa = 9.24KK372 pKa = 10.21RR373 pKa = 11.84DD374 pKa = 3.22RR375 pKa = 11.84NTASCINRR383 pKa = 11.84LCEE386 pKa = 3.97LMCEE390 pKa = 4.22VRR392 pKa = 11.84ALGQSPEE399 pKa = 4.03DD400 pKa = 3.35LLFVVEE406 pKa = 4.62WGGQIDD412 pKa = 4.22SAAVPAFCAMARR424 pKa = 11.84VDD426 pKa = 3.63CAFDD430 pKa = 3.44VGASGVDD437 pKa = 2.97IGGVADD443 pKa = 4.12PGKK446 pKa = 10.3SLEE449 pKa = 4.19VHH451 pKa = 6.87HH452 pKa = 5.79YY453 pKa = 8.72TAVLTHH459 pKa = 6.27RR460 pKa = 11.84VGRR463 pKa = 11.84TLSVCCEE470 pKa = 3.58VPYY473 pKa = 11.13AKK475 pKa = 10.46DD476 pKa = 2.84SGVNDD481 pKa = 5.17RR482 pKa = 11.84IRR484 pKa = 11.84TVVDD488 pKa = 3.44AVGEE492 pKa = 4.32GVKK495 pKa = 10.8GFVYY499 pKa = 10.66VSGGIPAVASMSAEE513 pKa = 3.66QSVNDD518 pKa = 3.82AHH520 pKa = 7.93SDD522 pKa = 2.94IYY524 pKa = 11.23AALVGDD530 pKa = 4.83APLKK534 pKa = 10.79CILFSTDD541 pKa = 4.56LIIPPACPHH550 pKa = 6.78CEE552 pKa = 4.06TVDD555 pKa = 3.81LDD557 pKa = 3.61HH558 pKa = 7.02FLASRR563 pKa = 11.84GVLSEE568 pKa = 4.14DD569 pKa = 3.48CRR571 pKa = 11.84EE572 pKa = 4.02CTVFGRR578 pKa = 11.84ALRR581 pKa = 11.84LMSEE585 pKa = 4.45LMSTPGVRR593 pKa = 11.84LVKK596 pKa = 9.68TRR598 pKa = 11.84SAFAHH603 pKa = 6.02NSQFDD608 pKa = 3.97LEE610 pKa = 4.43YY611 pKa = 11.14SSLLRR616 pKa = 11.84NTLTDD621 pKa = 2.84SSSAVDD627 pKa = 3.36SAAFSTGARR636 pKa = 11.84NRR638 pKa = 11.84TWGGGMPVGSDD649 pKa = 3.62DD650 pKa = 4.97LVAAQAIGGTVDD662 pKa = 3.16GEE664 pKa = 4.48FYY666 pKa = 11.24DD667 pKa = 3.74RR668 pKa = 11.84MVGIRR673 pKa = 11.84RR674 pKa = 11.84NVYY677 pKa = 10.19ALLAGGAQPRR687 pKa = 11.84PALADD692 pKa = 3.38DD693 pKa = 3.96TFSDD697 pKa = 4.09VVRR700 pKa = 11.84SVGAA704 pKa = 3.55
MM1 pKa = 7.89ADD3 pKa = 3.79LTRR6 pKa = 11.84LSAIAVDD13 pKa = 4.05VGSPAPAVAAVLDD26 pKa = 4.18LFGVSDD32 pKa = 3.81PTGQCLSSAQVMSVVEE48 pKa = 4.73WIAGFPLQSAPGLMPAYY65 pKa = 9.55VFEE68 pKa = 4.37YY69 pKa = 10.21VKK71 pKa = 10.92SKK73 pKa = 10.86AGSLVASAGVCDD85 pKa = 4.07DD86 pKa = 2.96VAYY89 pKa = 10.26FLRR92 pKa = 11.84HH93 pKa = 5.74FEE95 pKa = 4.87APDD98 pKa = 3.1EE99 pKa = 4.9HH100 pKa = 6.07YY101 pKa = 9.28TKK103 pKa = 10.22SGRR106 pKa = 11.84SARR109 pKa = 11.84VAQVALNKK117 pKa = 9.41PLKK120 pKa = 10.61GDD122 pKa = 3.81GAHH125 pKa = 6.94LGAVEE130 pKa = 4.76LALLDD135 pKa = 3.56AVTRR139 pKa = 11.84VGYY142 pKa = 9.93VGPAGALASVARR154 pKa = 11.84LRR156 pKa = 11.84TMEE159 pKa = 4.11VLRR162 pKa = 11.84HH163 pKa = 4.85YY164 pKa = 10.95AVDD167 pKa = 3.42VRR169 pKa = 11.84QRR171 pKa = 11.84ACAQYY176 pKa = 10.67RR177 pKa = 11.84VTCQVCLPGRR187 pKa = 11.84DD188 pKa = 3.82SYY190 pKa = 11.84PLITPWCVSRR200 pKa = 11.84QSATICAGVILASVGGEE217 pKa = 3.64AATKK221 pKa = 9.61AVQRR225 pKa = 11.84RR226 pKa = 11.84AVTVASHH233 pKa = 7.06AARR236 pKa = 11.84IGLDD240 pKa = 3.1AVRR243 pKa = 11.84AIVAPACRR251 pKa = 11.84LVAEE255 pKa = 4.44YY256 pKa = 11.03SFDD259 pKa = 3.68PVRR262 pKa = 11.84LVLTDD267 pKa = 3.5RR268 pKa = 11.84YY269 pKa = 10.28GRR271 pKa = 11.84VRR273 pKa = 11.84EE274 pKa = 4.38RR275 pKa = 11.84ITSRR279 pKa = 11.84SPAATVAFAYY289 pKa = 7.65LTSRR293 pKa = 11.84GSADD297 pKa = 3.79PLLAMASEE305 pKa = 4.56RR306 pKa = 11.84VEE308 pKa = 4.08RR309 pKa = 11.84MGPVEE314 pKa = 5.63LLPQCVQDD322 pKa = 3.3YY323 pKa = 9.09WNQSEE328 pKa = 5.53DD329 pKa = 3.49IVGMLSLFCRR339 pKa = 11.84ALHH342 pKa = 5.17LTRR345 pKa = 11.84VEE347 pKa = 4.01RR348 pKa = 11.84EE349 pKa = 3.68EE350 pKa = 4.05EE351 pKa = 3.85EE352 pKa = 4.31RR353 pKa = 11.84GRR355 pKa = 11.84DD356 pKa = 3.77VNKK359 pKa = 10.41GPCLGYY365 pKa = 10.38LAVGRR370 pKa = 11.84KK371 pKa = 9.24KK372 pKa = 10.21RR373 pKa = 11.84DD374 pKa = 3.22RR375 pKa = 11.84NTASCINRR383 pKa = 11.84LCEE386 pKa = 3.97LMCEE390 pKa = 4.22VRR392 pKa = 11.84ALGQSPEE399 pKa = 4.03DD400 pKa = 3.35LLFVVEE406 pKa = 4.62WGGQIDD412 pKa = 4.22SAAVPAFCAMARR424 pKa = 11.84VDD426 pKa = 3.63CAFDD430 pKa = 3.44VGASGVDD437 pKa = 2.97IGGVADD443 pKa = 4.12PGKK446 pKa = 10.3SLEE449 pKa = 4.19VHH451 pKa = 6.87HH452 pKa = 5.79YY453 pKa = 8.72TAVLTHH459 pKa = 6.27RR460 pKa = 11.84VGRR463 pKa = 11.84TLSVCCEE470 pKa = 3.58VPYY473 pKa = 11.13AKK475 pKa = 10.46DD476 pKa = 2.84SGVNDD481 pKa = 5.17RR482 pKa = 11.84IRR484 pKa = 11.84TVVDD488 pKa = 3.44AVGEE492 pKa = 4.32GVKK495 pKa = 10.8GFVYY499 pKa = 10.66VSGGIPAVASMSAEE513 pKa = 3.66QSVNDD518 pKa = 3.82AHH520 pKa = 7.93SDD522 pKa = 2.94IYY524 pKa = 11.23AALVGDD530 pKa = 4.83APLKK534 pKa = 10.79CILFSTDD541 pKa = 4.56LIIPPACPHH550 pKa = 6.78CEE552 pKa = 4.06TVDD555 pKa = 3.81LDD557 pKa = 3.61HH558 pKa = 7.02FLASRR563 pKa = 11.84GVLSEE568 pKa = 4.14DD569 pKa = 3.48CRR571 pKa = 11.84EE572 pKa = 4.02CTVFGRR578 pKa = 11.84ALRR581 pKa = 11.84LMSEE585 pKa = 4.45LMSTPGVRR593 pKa = 11.84LVKK596 pKa = 9.68TRR598 pKa = 11.84SAFAHH603 pKa = 6.02NSQFDD608 pKa = 3.97LEE610 pKa = 4.43YY611 pKa = 11.14SSLLRR616 pKa = 11.84NTLTDD621 pKa = 2.84SSSAVDD627 pKa = 3.36SAAFSTGARR636 pKa = 11.84NRR638 pKa = 11.84TWGGGMPVGSDD649 pKa = 3.62DD650 pKa = 4.97LVAAQAIGGTVDD662 pKa = 3.16GEE664 pKa = 4.48FYY666 pKa = 11.24DD667 pKa = 3.74RR668 pKa = 11.84MVGIRR673 pKa = 11.84RR674 pKa = 11.84NVYY677 pKa = 10.19ALLAGGAQPRR687 pKa = 11.84PALADD692 pKa = 3.38DD693 pKa = 3.96TFSDD697 pKa = 4.09VVRR700 pKa = 11.84SVGAA704 pKa = 3.55
Molecular weight: 74.96 kDa
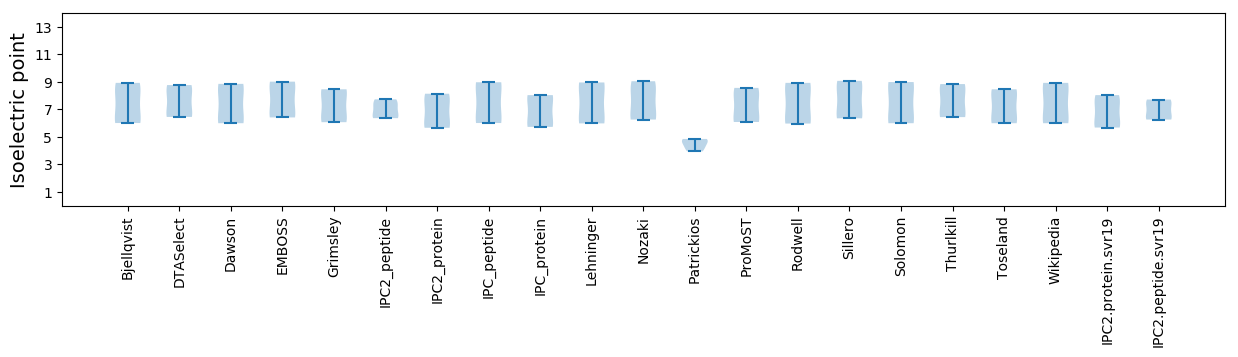
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1V1IEY9|A0A1V1IEY9_9VIRU RNA dependent RNA polymerase OS=Beauveria bassiana polymycovirus 1 OX=1740646 GN=RdRp PE=4 SV=1
MM1 pKa = 7.24SVSTVTSEE9 pKa = 3.78IAHH12 pKa = 5.31VQVACRR18 pKa = 11.84PVSEE22 pKa = 4.65MAPHH26 pKa = 6.9LQRR29 pKa = 11.84ASQDD33 pKa = 3.57TVQGALKK40 pKa = 9.67WVADD44 pKa = 3.88KK45 pKa = 11.14YY46 pKa = 10.93SRR48 pKa = 11.84GGVRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84NLTVHH59 pKa = 6.54LSGVRR64 pKa = 11.84GRR66 pKa = 11.84ADD68 pKa = 3.45DD69 pKa = 3.78VVLIPSVIEE78 pKa = 4.08ANSVLPEE85 pKa = 4.33APVHH89 pKa = 5.77RR90 pKa = 11.84APYY93 pKa = 10.07VDD95 pKa = 3.88IGGDD99 pKa = 3.65SADD102 pKa = 3.39SRR104 pKa = 11.84YY105 pKa = 10.6EE106 pKa = 3.9GLKK109 pKa = 11.06GLDD112 pKa = 2.93KK113 pKa = 11.06VQNKK117 pKa = 7.83GAKK120 pKa = 9.65GGAEE124 pKa = 4.24FEE126 pKa = 4.54RR127 pKa = 11.84KK128 pKa = 8.72SSRR131 pKa = 11.84MEE133 pKa = 3.4GAIRR137 pKa = 11.84KK138 pKa = 9.45RR139 pKa = 11.84GMLRR143 pKa = 11.84NPSFQVCRR151 pKa = 11.84YY152 pKa = 7.8QLPFSYY158 pKa = 9.95GYY160 pKa = 9.63KK161 pKa = 9.77ARR163 pKa = 11.84RR164 pKa = 11.84VTTEE168 pKa = 3.32RR169 pKa = 11.84HH170 pKa = 5.28LLGPTLDD177 pKa = 3.65ALHH180 pKa = 6.45RR181 pKa = 11.84AKK183 pKa = 10.43RR184 pKa = 11.84RR185 pKa = 11.84PGYY188 pKa = 8.54PQVVKK193 pKa = 10.91DD194 pKa = 4.21SLDD197 pKa = 3.55EE198 pKa = 4.31AVDD201 pKa = 3.71LGDD204 pKa = 4.04YY205 pKa = 7.41TTHH208 pKa = 7.86DD209 pKa = 4.61PDD211 pKa = 3.88TLHH214 pKa = 7.07KK215 pKa = 10.59RR216 pKa = 11.84FLEE219 pKa = 4.4YY220 pKa = 9.88VHH222 pKa = 6.52EE223 pKa = 4.29RR224 pKa = 11.84VEE226 pKa = 4.57GVSADD231 pKa = 3.46TKK233 pKa = 10.7TDD235 pKa = 3.57FLVAVQVMWRR245 pKa = 11.84VWSDD249 pKa = 2.85AGVAAEE255 pKa = 4.81ARR257 pKa = 11.84PFSDD261 pKa = 3.57ATPEE265 pKa = 4.1GLDD268 pKa = 4.15AMFTTGNAGEE278 pKa = 4.07YY279 pKa = 9.0RR280 pKa = 11.84KK281 pKa = 10.3HH282 pKa = 5.89GVEE285 pKa = 4.0SRR287 pKa = 11.84RR288 pKa = 11.84DD289 pKa = 3.48PRR291 pKa = 11.84ALDD294 pKa = 3.42MLSKK298 pKa = 10.13HH299 pKa = 5.07VQGFTVAGRR308 pKa = 11.84NMPTGMSPPAFVDD321 pKa = 3.4RR322 pKa = 11.84LDD324 pKa = 3.9HH325 pKa = 5.66VTTSFGKK332 pKa = 10.4RR333 pKa = 11.84EE334 pKa = 4.0PKK336 pKa = 9.37KK337 pKa = 10.55AKK339 pKa = 9.36IVNGQRR345 pKa = 11.84VAPVPRR351 pKa = 11.84FIFNPSPCSYY361 pKa = 11.68AMGAFLHH368 pKa = 6.68SDD370 pKa = 3.1ISHH373 pKa = 7.27ALQDD377 pKa = 3.95RR378 pKa = 11.84DD379 pKa = 3.88PLHH382 pKa = 6.87GPGFGPGRR390 pKa = 11.84GRR392 pKa = 11.84AVKK395 pKa = 8.62FTRR398 pKa = 11.84IVEE401 pKa = 4.19AACPDD406 pKa = 3.56GSRR409 pKa = 11.84LRR411 pKa = 11.84GGCKK415 pKa = 9.86AVMSDD420 pKa = 3.81IEE422 pKa = 4.16KK423 pKa = 9.89WDD425 pKa = 3.74ANMSEE430 pKa = 4.26FLIGTSFDD438 pKa = 3.46ALEE441 pKa = 4.33AFVDD445 pKa = 3.94KK446 pKa = 11.26SKK448 pKa = 10.79LAPLDD453 pKa = 3.47RR454 pKa = 11.84ASRR457 pKa = 11.84EE458 pKa = 3.73ASCRR462 pKa = 11.84YY463 pKa = 7.43MRR465 pKa = 11.84RR466 pKa = 11.84TLMEE470 pKa = 4.42KK471 pKa = 10.29LVEE474 pKa = 4.41HH475 pKa = 6.74PSGYY479 pKa = 10.2LVHH482 pKa = 7.29LYY484 pKa = 10.7GCMPSGSFYY493 pKa = 10.81TSLLNTVANTLLALALLARR512 pKa = 11.84RR513 pKa = 11.84MRR515 pKa = 11.84LAEE518 pKa = 4.07KK519 pKa = 9.96PIDD522 pKa = 3.45IAAMARR528 pKa = 11.84SADD531 pKa = 3.75GALLSYY537 pKa = 11.04GDD539 pKa = 3.8NQLIITSLFEE549 pKa = 3.97AHH551 pKa = 6.01GVEE554 pKa = 5.07YY555 pKa = 11.16DD556 pKa = 3.56MDD558 pKa = 3.85DD559 pKa = 3.48HH560 pKa = 8.55AEE562 pKa = 4.07LLSEE566 pKa = 4.16VGMRR570 pKa = 11.84LKK572 pKa = 10.52KK573 pKa = 10.73DD574 pKa = 3.23EE575 pKa = 4.62TEE577 pKa = 3.95VSDD580 pKa = 3.56RR581 pKa = 11.84LDD583 pKa = 3.92RR584 pKa = 11.84IRR586 pKa = 11.84FCSRR590 pKa = 11.84AVVRR594 pKa = 11.84TPAGMAITRR603 pKa = 11.84PHH605 pKa = 6.62GDD607 pKa = 3.07VVAKK611 pKa = 10.17LVARR615 pKa = 11.84PTEE618 pKa = 3.76NSVDD622 pKa = 3.44DD623 pKa = 4.17KK624 pKa = 11.6LYY626 pKa = 11.07VRR628 pKa = 11.84ALMADD633 pKa = 3.45YY634 pKa = 10.18MGVDD638 pKa = 4.52PIVHH642 pKa = 7.12RR643 pKa = 11.84ILADD647 pKa = 3.08VDD649 pKa = 3.15ATIRR653 pKa = 11.84VTIDD657 pKa = 2.81AVRR660 pKa = 11.84ASEE663 pKa = 4.04RR664 pKa = 11.84HH665 pKa = 5.91RR666 pKa = 11.84DD667 pKa = 3.77TLRR670 pKa = 11.84SAASQVFGRR679 pKa = 11.84KK680 pKa = 9.5DD681 pKa = 3.31DD682 pKa = 4.18DD683 pKa = 5.39AIAATAQLMAKK694 pKa = 9.97AVIDD698 pKa = 3.64RR699 pKa = 11.84RR700 pKa = 11.84VLLEE704 pKa = 3.68LTQPRR709 pKa = 11.84SVNDD713 pKa = 3.06VDD715 pKa = 4.98SVLEE719 pKa = 3.99RR720 pKa = 11.84TGKK723 pKa = 7.83VTGVRR728 pKa = 11.84FRR730 pKa = 11.84QRR732 pKa = 11.84FAASVGIQTDD742 pKa = 3.94VPSLVGPAAWLASQDD757 pKa = 3.41EE758 pKa = 4.33QQYY761 pKa = 11.14LDD763 pKa = 3.59YY764 pKa = 10.88LVRR767 pKa = 11.84TDD769 pKa = 3.46QVGVMHH775 pKa = 7.4
MM1 pKa = 7.24SVSTVTSEE9 pKa = 3.78IAHH12 pKa = 5.31VQVACRR18 pKa = 11.84PVSEE22 pKa = 4.65MAPHH26 pKa = 6.9LQRR29 pKa = 11.84ASQDD33 pKa = 3.57TVQGALKK40 pKa = 9.67WVADD44 pKa = 3.88KK45 pKa = 11.14YY46 pKa = 10.93SRR48 pKa = 11.84GGVRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84NLTVHH59 pKa = 6.54LSGVRR64 pKa = 11.84GRR66 pKa = 11.84ADD68 pKa = 3.45DD69 pKa = 3.78VVLIPSVIEE78 pKa = 4.08ANSVLPEE85 pKa = 4.33APVHH89 pKa = 5.77RR90 pKa = 11.84APYY93 pKa = 10.07VDD95 pKa = 3.88IGGDD99 pKa = 3.65SADD102 pKa = 3.39SRR104 pKa = 11.84YY105 pKa = 10.6EE106 pKa = 3.9GLKK109 pKa = 11.06GLDD112 pKa = 2.93KK113 pKa = 11.06VQNKK117 pKa = 7.83GAKK120 pKa = 9.65GGAEE124 pKa = 4.24FEE126 pKa = 4.54RR127 pKa = 11.84KK128 pKa = 8.72SSRR131 pKa = 11.84MEE133 pKa = 3.4GAIRR137 pKa = 11.84KK138 pKa = 9.45RR139 pKa = 11.84GMLRR143 pKa = 11.84NPSFQVCRR151 pKa = 11.84YY152 pKa = 7.8QLPFSYY158 pKa = 9.95GYY160 pKa = 9.63KK161 pKa = 9.77ARR163 pKa = 11.84RR164 pKa = 11.84VTTEE168 pKa = 3.32RR169 pKa = 11.84HH170 pKa = 5.28LLGPTLDD177 pKa = 3.65ALHH180 pKa = 6.45RR181 pKa = 11.84AKK183 pKa = 10.43RR184 pKa = 11.84RR185 pKa = 11.84PGYY188 pKa = 8.54PQVVKK193 pKa = 10.91DD194 pKa = 4.21SLDD197 pKa = 3.55EE198 pKa = 4.31AVDD201 pKa = 3.71LGDD204 pKa = 4.04YY205 pKa = 7.41TTHH208 pKa = 7.86DD209 pKa = 4.61PDD211 pKa = 3.88TLHH214 pKa = 7.07KK215 pKa = 10.59RR216 pKa = 11.84FLEE219 pKa = 4.4YY220 pKa = 9.88VHH222 pKa = 6.52EE223 pKa = 4.29RR224 pKa = 11.84VEE226 pKa = 4.57GVSADD231 pKa = 3.46TKK233 pKa = 10.7TDD235 pKa = 3.57FLVAVQVMWRR245 pKa = 11.84VWSDD249 pKa = 2.85AGVAAEE255 pKa = 4.81ARR257 pKa = 11.84PFSDD261 pKa = 3.57ATPEE265 pKa = 4.1GLDD268 pKa = 4.15AMFTTGNAGEE278 pKa = 4.07YY279 pKa = 9.0RR280 pKa = 11.84KK281 pKa = 10.3HH282 pKa = 5.89GVEE285 pKa = 4.0SRR287 pKa = 11.84RR288 pKa = 11.84DD289 pKa = 3.48PRR291 pKa = 11.84ALDD294 pKa = 3.42MLSKK298 pKa = 10.13HH299 pKa = 5.07VQGFTVAGRR308 pKa = 11.84NMPTGMSPPAFVDD321 pKa = 3.4RR322 pKa = 11.84LDD324 pKa = 3.9HH325 pKa = 5.66VTTSFGKK332 pKa = 10.4RR333 pKa = 11.84EE334 pKa = 4.0PKK336 pKa = 9.37KK337 pKa = 10.55AKK339 pKa = 9.36IVNGQRR345 pKa = 11.84VAPVPRR351 pKa = 11.84FIFNPSPCSYY361 pKa = 11.68AMGAFLHH368 pKa = 6.68SDD370 pKa = 3.1ISHH373 pKa = 7.27ALQDD377 pKa = 3.95RR378 pKa = 11.84DD379 pKa = 3.88PLHH382 pKa = 6.87GPGFGPGRR390 pKa = 11.84GRR392 pKa = 11.84AVKK395 pKa = 8.62FTRR398 pKa = 11.84IVEE401 pKa = 4.19AACPDD406 pKa = 3.56GSRR409 pKa = 11.84LRR411 pKa = 11.84GGCKK415 pKa = 9.86AVMSDD420 pKa = 3.81IEE422 pKa = 4.16KK423 pKa = 9.89WDD425 pKa = 3.74ANMSEE430 pKa = 4.26FLIGTSFDD438 pKa = 3.46ALEE441 pKa = 4.33AFVDD445 pKa = 3.94KK446 pKa = 11.26SKK448 pKa = 10.79LAPLDD453 pKa = 3.47RR454 pKa = 11.84ASRR457 pKa = 11.84EE458 pKa = 3.73ASCRR462 pKa = 11.84YY463 pKa = 7.43MRR465 pKa = 11.84RR466 pKa = 11.84TLMEE470 pKa = 4.42KK471 pKa = 10.29LVEE474 pKa = 4.41HH475 pKa = 6.74PSGYY479 pKa = 10.2LVHH482 pKa = 7.29LYY484 pKa = 10.7GCMPSGSFYY493 pKa = 10.81TSLLNTVANTLLALALLARR512 pKa = 11.84RR513 pKa = 11.84MRR515 pKa = 11.84LAEE518 pKa = 4.07KK519 pKa = 9.96PIDD522 pKa = 3.45IAAMARR528 pKa = 11.84SADD531 pKa = 3.75GALLSYY537 pKa = 11.04GDD539 pKa = 3.8NQLIITSLFEE549 pKa = 3.97AHH551 pKa = 6.01GVEE554 pKa = 5.07YY555 pKa = 11.16DD556 pKa = 3.56MDD558 pKa = 3.85DD559 pKa = 3.48HH560 pKa = 8.55AEE562 pKa = 4.07LLSEE566 pKa = 4.16VGMRR570 pKa = 11.84LKK572 pKa = 10.52KK573 pKa = 10.73DD574 pKa = 3.23EE575 pKa = 4.62TEE577 pKa = 3.95VSDD580 pKa = 3.56RR581 pKa = 11.84LDD583 pKa = 3.92RR584 pKa = 11.84IRR586 pKa = 11.84FCSRR590 pKa = 11.84AVVRR594 pKa = 11.84TPAGMAITRR603 pKa = 11.84PHH605 pKa = 6.62GDD607 pKa = 3.07VVAKK611 pKa = 10.17LVARR615 pKa = 11.84PTEE618 pKa = 3.76NSVDD622 pKa = 3.44DD623 pKa = 4.17KK624 pKa = 11.6LYY626 pKa = 11.07VRR628 pKa = 11.84ALMADD633 pKa = 3.45YY634 pKa = 10.18MGVDD638 pKa = 4.52PIVHH642 pKa = 7.12RR643 pKa = 11.84ILADD647 pKa = 3.08VDD649 pKa = 3.15ATIRR653 pKa = 11.84VTIDD657 pKa = 2.81AVRR660 pKa = 11.84ASEE663 pKa = 4.04RR664 pKa = 11.84HH665 pKa = 5.91RR666 pKa = 11.84DD667 pKa = 3.77TLRR670 pKa = 11.84SAASQVFGRR679 pKa = 11.84KK680 pKa = 9.5DD681 pKa = 3.31DD682 pKa = 4.18DD683 pKa = 5.39AIAATAQLMAKK694 pKa = 9.97AVIDD698 pKa = 3.64RR699 pKa = 11.84RR700 pKa = 11.84VLLEE704 pKa = 3.68LTQPRR709 pKa = 11.84SVNDD713 pKa = 3.06VDD715 pKa = 4.98SVLEE719 pKa = 3.99RR720 pKa = 11.84TGKK723 pKa = 7.83VTGVRR728 pKa = 11.84FRR730 pKa = 11.84QRR732 pKa = 11.84FAASVGIQTDD742 pKa = 3.94VPSLVGPAAWLASQDD757 pKa = 3.41EE758 pKa = 4.33QQYY761 pKa = 11.14LDD763 pKa = 3.59YY764 pKa = 10.88LVRR767 pKa = 11.84TDD769 pKa = 3.46QVGVMHH775 pKa = 7.4
Molecular weight: 85.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2395 |
306 |
775 |
598.8 |
64.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.608 ± 1.086 | 1.879 ± 0.549 |
7.516 ± 0.407 | 4.635 ± 0.05 |
3.215 ± 0.432 | 8.058 ± 0.505 |
2.255 ± 0.345 | 3.006 ± 0.104 |
3.048 ± 0.562 | 8.225 ± 0.39 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.296 ± 0.3 | 1.461 ± 0.168 |
5.136 ± 0.335 | 2.756 ± 0.164 |
8.894 ± 0.354 | 7.432 ± 0.237 |
4.676 ± 0.19 | 10.647 ± 0.857 |
0.752 ± 0.052 | 2.505 ± 0.213 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
