
Ectothiorhodospiraceae bacterium BW-2
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Ectothiorhodospiraceae; unclassified Ectothiorhodospiraceae
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
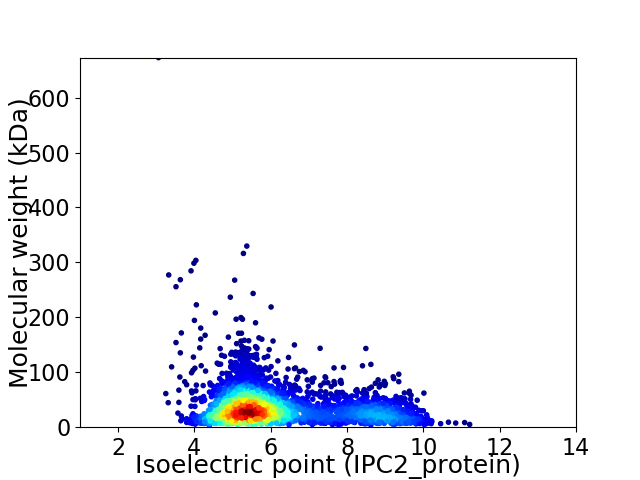
Virtual 2D-PAGE plot for 3445 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C2HU83|A0A5C2HU83_9GAMM Methyl-accepting chemotaxis protein OS=Ectothiorhodospiraceae bacterium BW-2 OX=947515 GN=D5085_15465 PE=4 SV=1
MM1 pKa = 6.61TQVTATTDD9 pKa = 3.15SSSVSFDD16 pKa = 3.03EE17 pKa = 4.26TAPALTAVSIASDD30 pKa = 3.51NSDD33 pKa = 3.06TTLAKK38 pKa = 10.13TGDD41 pKa = 3.74TVTLSFTTDD50 pKa = 3.17GSHH53 pKa = 6.86SGTPTVTLGGNGVTATNTSGNTYY76 pKa = 7.73TASYY80 pKa = 8.85TLQAGDD86 pKa = 3.5TEE88 pKa = 4.82GAVSFTIDD96 pKa = 3.36AVDD99 pKa = 3.24AAGNAMTQVTATTDD113 pKa = 3.15SSSVSFDD120 pKa = 3.03EE121 pKa = 4.26TAPALTAVSIASDD134 pKa = 3.5NSDD137 pKa = 3.11TALAKK142 pKa = 10.19TGDD145 pKa = 3.77TVTLSFTTDD154 pKa = 3.17GSHH157 pKa = 6.86SGTPTVTLGGNSVTATNTSGNTYY180 pKa = 7.73TASYY184 pKa = 8.85TLQAGDD190 pKa = 3.5TEE192 pKa = 4.82GAVSFTIDD200 pKa = 3.36AVDD203 pKa = 3.24AAGNAMTQVTATTDD217 pKa = 3.15SSSVSFDD224 pKa = 3.03EE225 pKa = 4.26TAPALTAVSIASDD238 pKa = 3.51NSDD241 pKa = 3.08TTLAKK246 pKa = 10.86VGDD249 pKa = 4.21TVTLSFTTDD258 pKa = 3.17GSHH261 pKa = 6.86SGTPTVTLGGNGVTATNTSGNTYY284 pKa = 7.73TASYY288 pKa = 8.85TLQAGDD294 pKa = 3.5TEE296 pKa = 4.82GAVSFTIDD304 pKa = 3.36AVDD307 pKa = 3.24AAGNAMTQVTATTDD321 pKa = 3.15SSSVSFDD328 pKa = 3.03EE329 pKa = 4.26TAPALTAVSIASDD342 pKa = 3.51NSDD345 pKa = 3.08TTLAKK350 pKa = 10.86VGDD353 pKa = 4.21TVTLSFTTDD362 pKa = 3.17GSHH365 pKa = 6.86SGTPTVTLGGNSVTATNTSGNTYY388 pKa = 7.73TASYY392 pKa = 8.74TLQAGDD398 pKa = 3.61IEE400 pKa = 4.76GTLSFTIDD408 pKa = 3.19AVDD411 pKa = 3.24AAGNAMTQVTATTDD425 pKa = 3.15SSSVSFDD432 pKa = 3.03EE433 pKa = 4.26TAPALTAVSIASDD446 pKa = 3.51NSDD449 pKa = 3.08TTLAKK454 pKa = 10.86VGDD457 pKa = 4.21TVTLSFTTDD466 pKa = 3.17GSHH469 pKa = 6.86SGTPTVTLGGNSVTATNTSGNTYY492 pKa = 7.73TASYY496 pKa = 8.85TLQAGDD502 pKa = 3.5TEE504 pKa = 4.82GAVSFTIDD512 pKa = 3.36AVDD515 pKa = 3.24AAGNAMTQVTATTDD529 pKa = 3.15SSSVSFDD536 pKa = 3.03TTAPALTAVSIASDD550 pKa = 3.5NSDD553 pKa = 3.11TALAKK558 pKa = 10.19TGDD561 pKa = 3.77TVTLSFTTDD570 pKa = 3.17GSHH573 pKa = 6.86SGTPTVTLGGNGVTATNTSGNTYY596 pKa = 7.73TASYY600 pKa = 8.85TLQAGDD606 pKa = 3.5TEE608 pKa = 4.86GAVSFSIDD616 pKa = 3.24AVDD619 pKa = 3.4AAGNAMTQVTATTDD633 pKa = 3.15SSSVSFDD640 pKa = 3.03EE641 pKa = 4.26TAPALTAVSIASDD654 pKa = 3.51NSDD657 pKa = 3.08TTLAKK662 pKa = 10.86VGDD665 pKa = 4.21TVTLSFTTDD674 pKa = 3.17GSHH677 pKa = 6.86SGTPTVTLGGNSVTATNTSGNTYY700 pKa = 7.73TASYY704 pKa = 8.85TLQAGDD710 pKa = 3.5TEE712 pKa = 4.82GAVSFTIDD720 pKa = 3.36AVDD723 pKa = 3.24AAGNAMTQVTATTDD737 pKa = 3.15SSSVSFDD744 pKa = 3.03EE745 pKa = 4.17TAPTINSVSVADD757 pKa = 3.88GTYY760 pKa = 10.38KK761 pKa = 10.36IGDD764 pKa = 4.63AITITVTAGSNEE776 pKa = 4.2TGLTLAGTFNGQTLTGITDD795 pKa = 3.84NNDD798 pKa = 2.67GTYY801 pKa = 10.42SATYY805 pKa = 8.46TAVEE809 pKa = 4.29GDD811 pKa = 3.83SDD813 pKa = 4.02VADD816 pKa = 4.31AGSVTTNITLTDD828 pKa = 3.48AAGNASSATTSVTLAGEE845 pKa = 4.56SIDD848 pKa = 4.09GNSPTGSVSDD858 pKa = 4.19LVTFPTIQLEE868 pKa = 4.32ATGYY872 pKa = 5.74TTGEE876 pKa = 3.85DD877 pKa = 3.05RR878 pKa = 11.84YY879 pKa = 10.9VALEE883 pKa = 4.09SVGSDD888 pKa = 2.61GGYY891 pKa = 8.51VVAWEE896 pKa = 4.71GEE898 pKa = 4.28YY899 pKa = 11.1GSYY902 pKa = 9.47PLKK905 pKa = 10.6SIYY908 pKa = 9.67VQQFNADD915 pKa = 3.57GTTTGYY921 pKa = 10.95SPVQLDD927 pKa = 4.51GIGVTDD933 pKa = 3.76SYY935 pKa = 11.78DD936 pKa = 3.51YY937 pKa = 10.71RR938 pKa = 11.84PKK940 pKa = 9.49ITALEE945 pKa = 4.1SGGYY949 pKa = 8.53VVTWSGLDD957 pKa = 3.45DD958 pKa = 4.97SGDD961 pKa = 3.41NDD963 pKa = 3.52TSVFVQQFNADD974 pKa = 3.72GTTTGYY980 pKa = 11.28DD981 pKa = 3.6PVQLEE986 pKa = 4.28ATGYY990 pKa = 7.46TAGEE994 pKa = 4.17DD995 pKa = 3.97DD996 pKa = 4.35EE997 pKa = 4.75PQITSLGSSGEE1008 pKa = 4.04YY1009 pKa = 9.08VVIWRR1014 pKa = 11.84GQNSSTSSDD1023 pKa = 2.65RR1024 pKa = 11.84SIFVQQFNANGTVKK1038 pKa = 10.7NDD1040 pKa = 3.35TPVILEE1046 pKa = 4.08AHH1048 pKa = 6.66GSWDD1052 pKa = 3.3QYY1054 pKa = 9.09QQITTLNSGGYY1065 pKa = 8.41VVTWTGLDD1073 pKa = 3.3GTIYY1077 pKa = 10.4PDD1079 pKa = 3.4TDD1081 pKa = 3.29YY1082 pKa = 11.37SIYY1085 pKa = 10.3VQQFNADD1092 pKa = 3.59GTRR1095 pKa = 11.84TGDD1098 pKa = 3.51TPVKK1102 pKa = 10.83LEE1104 pKa = 4.01GTGNTTGGDD1113 pKa = 3.2EE1114 pKa = 4.3HH1115 pKa = 8.04SQVTALDD1122 pKa = 3.78FNGGYY1127 pKa = 8.93VVAWKK1132 pKa = 9.61GQNSDD1137 pKa = 3.14NRR1139 pKa = 11.84DD1140 pKa = 3.7SIFVQQFDD1148 pKa = 4.1ANGTITGYY1156 pKa = 9.07TPVQLNSHH1164 pKa = 6.08SNDD1167 pKa = 2.97RR1168 pKa = 11.84DD1169 pKa = 3.64VQIAAVGSSGQYY1181 pKa = 7.72VATWVGMDD1189 pKa = 3.59SNYY1192 pKa = 10.72SYY1194 pKa = 11.18DD1195 pKa = 3.48IFVQQFNASGTTDD1208 pKa = 2.73GFTTVKK1214 pKa = 10.5LEE1216 pKa = 4.54SINNPDD1222 pKa = 3.55SGNGAKK1228 pKa = 10.12PKK1230 pKa = 10.06IAALGDD1236 pKa = 3.34SGRR1239 pKa = 11.84YY1240 pKa = 8.63LVTWYY1245 pKa = 10.11DD1246 pKa = 3.4YY1247 pKa = 11.46DD1248 pKa = 3.92EE1249 pKa = 5.44EE1250 pKa = 6.28GDD1252 pKa = 3.61EE1253 pKa = 5.6SIFLQAFNADD1263 pKa = 3.45GTTAGPTQQFEE1274 pKa = 4.52ATGNTTGSDD1283 pKa = 3.45QYY1285 pKa = 11.55PEE1287 pKa = 4.03VLALGDD1293 pKa = 3.32SGAFVLAWHH1302 pKa = 6.61GRR1304 pKa = 11.84DD1305 pKa = 3.3SDD1307 pKa = 4.13DD1308 pKa = 4.51DD1309 pKa = 4.1VSIFVKK1315 pKa = 10.54QFFIDD1320 pKa = 3.65GMVRR1324 pKa = 11.84AQSSEE1329 pKa = 3.64AATAYY1334 pKa = 10.07LVKK1337 pKa = 10.27DD1338 pKa = 4.23TITVNSVTDD1347 pKa = 3.43LTGAPDD1353 pKa = 3.47SQMNTVAITSANTDD1367 pKa = 2.67TDD1369 pKa = 4.03LPLTGLEE1376 pKa = 4.03EE1377 pKa = 4.54GNYY1380 pKa = 9.07TLYY1383 pKa = 11.16LVDD1386 pKa = 3.34AAGNIGTVGTSVTVDD1401 pKa = 3.32HH1402 pKa = 6.97TPPNAPTISAVTDD1415 pKa = 3.65DD1416 pKa = 3.93QGSVTGTVADD1426 pKa = 5.16DD1427 pKa = 5.12GISDD1431 pKa = 3.93DD1432 pKa = 4.14TILTVRR1438 pKa = 11.84VSLDD1442 pKa = 3.17STNALSGDD1450 pKa = 3.82SVQLYY1455 pKa = 10.77DD1456 pKa = 4.16GFSALGTAVTLTSTDD1471 pKa = 2.73ISNGYY1476 pKa = 9.6VDD1478 pKa = 5.05IDD1480 pKa = 4.28TPTLTAGTSYY1490 pKa = 11.25NFRR1493 pKa = 11.84ANITDD1498 pKa = 3.19AFGNIGEE1505 pKa = 4.54FSTDD1509 pKa = 2.66GAADD1513 pKa = 3.56VTIDD1517 pKa = 3.27TTAPDD1522 pKa = 3.97GNVVSTTAFVTLEE1535 pKa = 3.96ASGVTNMDD1543 pKa = 3.57DD1544 pKa = 4.1RR1545 pKa = 11.84NPQISAIGSNGEE1557 pKa = 4.38YY1558 pKa = 10.25VVTWTGGDD1566 pKa = 3.7GPDD1569 pKa = 5.09DD1570 pKa = 4.79YY1571 pKa = 11.74DD1572 pKa = 4.47DD1573 pKa = 4.77SIYY1576 pKa = 10.67VQHH1579 pKa = 6.9FNTDD1583 pKa = 2.96GTITGKK1589 pKa = 10.48AQVKK1593 pKa = 10.24LEE1595 pKa = 4.26STGVTDD1601 pKa = 5.17GGDD1604 pKa = 3.38SAPQVTSVGISGEE1617 pKa = 4.49YY1618 pKa = 8.94VVTWVGNDD1626 pKa = 3.55GPDD1629 pKa = 4.22DD1630 pKa = 4.74YY1631 pKa = 11.75DD1632 pKa = 4.4DD1633 pKa = 4.77SIYY1636 pKa = 10.63VQHH1639 pKa = 6.62FNADD1643 pKa = 3.27GTITGKK1649 pKa = 10.51AQVKK1653 pKa = 10.24LEE1655 pKa = 4.24STGVTDD1661 pKa = 5.59RR1662 pKa = 11.84GDD1664 pKa = 3.37EE1665 pKa = 4.25APQVTSVGISGEE1677 pKa = 4.49YY1678 pKa = 8.94VVTWVGNDD1686 pKa = 3.46GPGDD1690 pKa = 4.06YY1691 pKa = 10.52DD1692 pKa = 4.01DD1693 pKa = 5.56SIYY1696 pKa = 10.67VQHH1699 pKa = 6.62FNADD1703 pKa = 3.27GTITGKK1709 pKa = 10.47AQVKK1713 pKa = 9.97LEE1715 pKa = 4.14PTGVTDD1721 pKa = 6.01RR1722 pKa = 11.84GDD1724 pKa = 3.57KK1725 pKa = 10.71DD1726 pKa = 3.66PQVTSVGISGEE1737 pKa = 4.49YY1738 pKa = 8.94VVTWVGNDD1746 pKa = 3.43GSDD1749 pKa = 3.04EE1750 pKa = 4.42SIYY1753 pKa = 10.16VQHH1756 pKa = 6.6FNADD1760 pKa = 3.34GTITGKK1766 pKa = 10.27DD1767 pKa = 3.47QVKK1770 pKa = 10.64LEE1772 pKa = 4.19PTGQLSGDD1780 pKa = 3.82DD1781 pKa = 4.34LRR1783 pKa = 11.84PQVSAVGSNGEE1794 pKa = 4.28YY1795 pKa = 9.75VVVWDD1800 pKa = 5.26GVDD1803 pKa = 3.24ADD1805 pKa = 3.91GDD1807 pKa = 3.54RR1808 pKa = 11.84SVFVQRR1814 pKa = 11.84FNADD1818 pKa = 2.9GTKK1821 pKa = 10.64NGTEE1825 pKa = 4.08VLLEE1829 pKa = 4.01ATGVTDD1835 pKa = 4.78QSDD1838 pKa = 3.92MQAKK1842 pKa = 9.24IAAIGSSGEE1851 pKa = 4.67FVVTWPGKK1859 pKa = 10.41DD1860 pKa = 3.11SDD1862 pKa = 4.17GDD1864 pKa = 3.62DD1865 pKa = 4.04SIFVQHH1871 pKa = 6.36FNADD1875 pKa = 3.41GSTTNHH1881 pKa = 6.79SIVQLEE1887 pKa = 4.6AIGVTDD1893 pKa = 5.72DD1894 pKa = 4.46RR1895 pKa = 11.84DD1896 pKa = 3.71EE1897 pKa = 4.56NPQITALANSGYY1909 pKa = 10.06VVTWLGDD1916 pKa = 3.46DD1917 pKa = 4.54ANNDD1921 pKa = 3.53DD1922 pKa = 5.3QVYY1925 pKa = 9.39LQQFNADD1932 pKa = 3.54GSKK1935 pKa = 10.27SGSVTLVEE1943 pKa = 4.27VGNNEE1948 pKa = 4.09IASKK1952 pKa = 10.5GYY1954 pKa = 9.81PEE1956 pKa = 4.03IAAVGIGSQFAITSRR1971 pKa = 11.84GDD1973 pKa = 3.27DD1974 pKa = 3.25SGGDD1978 pKa = 3.21DD1979 pKa = 3.23SVYY1982 pKa = 10.55VRR1984 pKa = 11.84QFDD1987 pKa = 3.52ASTVFNNSKK1996 pKa = 8.37TVQVQSDD2003 pKa = 4.75GPGMAYY2009 pKa = 10.6LIKK2012 pKa = 10.75DD2013 pKa = 3.57SVTINSAADD2022 pKa = 3.55FSGVTDD2028 pKa = 3.58SQINSVSITSANTFTDD2044 pKa = 3.58LTLSGLEE2051 pKa = 3.9AGDD2054 pKa = 3.58YY2055 pKa = 10.82KK2056 pKa = 11.2LYY2058 pKa = 11.01LVDD2061 pKa = 3.53AAGNLGITTDD2071 pKa = 4.47GLTIDD2076 pKa = 4.0NTAPTASVTAATIGSSGNAVVQSTEE2101 pKa = 3.47AGTAFLVKK2109 pKa = 10.57DD2110 pKa = 4.18SITVNSLSDD2119 pKa = 3.13ITGASDD2125 pKa = 5.1NNWNWVTISTASSDD2139 pKa = 3.95TNLAAAGLVDD2149 pKa = 4.58GDD2151 pKa = 4.11YY2152 pKa = 11.05KK2153 pKa = 11.51VYY2155 pKa = 8.47TTDD2158 pKa = 3.17SAGNLSTVSTNSVTIDD2174 pKa = 3.35TTPPTTTYY2182 pKa = 10.91SGVSYY2187 pKa = 11.51DD2188 pKa = 3.6MASNTLTLTGTGINSMLEE2206 pKa = 4.07SGEE2209 pKa = 4.91DD2210 pKa = 3.15ATTDD2214 pKa = 3.08IKK2216 pKa = 11.45DD2217 pKa = 3.48RR2218 pKa = 11.84LDD2220 pKa = 3.07WSKK2223 pKa = 11.19LVWTFNNNTSYY2234 pKa = 11.61DD2235 pKa = 3.78FGSGDD2240 pKa = 3.78LGNNTGDD2247 pKa = 5.15DD2248 pKa = 3.74ILSVKK2253 pKa = 10.3AVSDD2257 pKa = 3.49TSLEE2261 pKa = 3.93ITFTDD2266 pKa = 4.53DD2267 pKa = 3.33AANACGSGLEE2277 pKa = 4.31SFFNGFDD2284 pKa = 3.46TSNPNYY2290 pKa = 10.74APFSDD2295 pKa = 3.65GHH2297 pKa = 6.42KK2298 pKa = 9.49LTVTAGFSRR2307 pKa = 11.84DD2308 pKa = 3.64SNGNAATGDD2317 pKa = 4.03GYY2319 pKa = 11.74VNPDD2323 pKa = 3.0TSIVVFDD2330 pKa = 5.32LIQGVSSSHH2339 pKa = 6.25SSRR2342 pKa = 11.84TFDD2345 pKa = 3.97ANTSYY2350 pKa = 10.76TIYY2353 pKa = 10.62IRR2355 pKa = 11.84MDD2357 pKa = 3.5SDD2359 pKa = 3.71SSTLSTAGSGPGTWGTWSSANNLGTDD2385 pKa = 3.62DD2386 pKa = 3.89KK2387 pKa = 11.64VIFVGSGSVIKK2398 pKa = 10.9NNSNPAQEE2406 pKa = 3.85VTFYY2410 pKa = 10.84YY2411 pKa = 10.55KK2412 pKa = 10.44SAGLLEE2418 pKa = 4.91LNYY2421 pKa = 10.39SLNSRR2426 pKa = 11.84TVQLYY2431 pKa = 9.05IYY2433 pKa = 9.7LGVGSIARR2441 pKa = 11.84NNYY2444 pKa = 9.63ASSEE2448 pKa = 4.17KK2449 pKa = 10.51LWDD2452 pKa = 3.77GNISVGDD2459 pKa = 4.03LRR2461 pKa = 11.84PTTYY2465 pKa = 9.72STGKK2469 pKa = 10.47FNDD2472 pKa = 3.93FVLTTMPNGILTSQGLVV2489 pKa = 3.12
MM1 pKa = 6.61TQVTATTDD9 pKa = 3.15SSSVSFDD16 pKa = 3.03EE17 pKa = 4.26TAPALTAVSIASDD30 pKa = 3.51NSDD33 pKa = 3.06TTLAKK38 pKa = 10.13TGDD41 pKa = 3.74TVTLSFTTDD50 pKa = 3.17GSHH53 pKa = 6.86SGTPTVTLGGNGVTATNTSGNTYY76 pKa = 7.73TASYY80 pKa = 8.85TLQAGDD86 pKa = 3.5TEE88 pKa = 4.82GAVSFTIDD96 pKa = 3.36AVDD99 pKa = 3.24AAGNAMTQVTATTDD113 pKa = 3.15SSSVSFDD120 pKa = 3.03EE121 pKa = 4.26TAPALTAVSIASDD134 pKa = 3.5NSDD137 pKa = 3.11TALAKK142 pKa = 10.19TGDD145 pKa = 3.77TVTLSFTTDD154 pKa = 3.17GSHH157 pKa = 6.86SGTPTVTLGGNSVTATNTSGNTYY180 pKa = 7.73TASYY184 pKa = 8.85TLQAGDD190 pKa = 3.5TEE192 pKa = 4.82GAVSFTIDD200 pKa = 3.36AVDD203 pKa = 3.24AAGNAMTQVTATTDD217 pKa = 3.15SSSVSFDD224 pKa = 3.03EE225 pKa = 4.26TAPALTAVSIASDD238 pKa = 3.51NSDD241 pKa = 3.08TTLAKK246 pKa = 10.86VGDD249 pKa = 4.21TVTLSFTTDD258 pKa = 3.17GSHH261 pKa = 6.86SGTPTVTLGGNGVTATNTSGNTYY284 pKa = 7.73TASYY288 pKa = 8.85TLQAGDD294 pKa = 3.5TEE296 pKa = 4.82GAVSFTIDD304 pKa = 3.36AVDD307 pKa = 3.24AAGNAMTQVTATTDD321 pKa = 3.15SSSVSFDD328 pKa = 3.03EE329 pKa = 4.26TAPALTAVSIASDD342 pKa = 3.51NSDD345 pKa = 3.08TTLAKK350 pKa = 10.86VGDD353 pKa = 4.21TVTLSFTTDD362 pKa = 3.17GSHH365 pKa = 6.86SGTPTVTLGGNSVTATNTSGNTYY388 pKa = 7.73TASYY392 pKa = 8.74TLQAGDD398 pKa = 3.61IEE400 pKa = 4.76GTLSFTIDD408 pKa = 3.19AVDD411 pKa = 3.24AAGNAMTQVTATTDD425 pKa = 3.15SSSVSFDD432 pKa = 3.03EE433 pKa = 4.26TAPALTAVSIASDD446 pKa = 3.51NSDD449 pKa = 3.08TTLAKK454 pKa = 10.86VGDD457 pKa = 4.21TVTLSFTTDD466 pKa = 3.17GSHH469 pKa = 6.86SGTPTVTLGGNSVTATNTSGNTYY492 pKa = 7.73TASYY496 pKa = 8.85TLQAGDD502 pKa = 3.5TEE504 pKa = 4.82GAVSFTIDD512 pKa = 3.36AVDD515 pKa = 3.24AAGNAMTQVTATTDD529 pKa = 3.15SSSVSFDD536 pKa = 3.03TTAPALTAVSIASDD550 pKa = 3.5NSDD553 pKa = 3.11TALAKK558 pKa = 10.19TGDD561 pKa = 3.77TVTLSFTTDD570 pKa = 3.17GSHH573 pKa = 6.86SGTPTVTLGGNGVTATNTSGNTYY596 pKa = 7.73TASYY600 pKa = 8.85TLQAGDD606 pKa = 3.5TEE608 pKa = 4.86GAVSFSIDD616 pKa = 3.24AVDD619 pKa = 3.4AAGNAMTQVTATTDD633 pKa = 3.15SSSVSFDD640 pKa = 3.03EE641 pKa = 4.26TAPALTAVSIASDD654 pKa = 3.51NSDD657 pKa = 3.08TTLAKK662 pKa = 10.86VGDD665 pKa = 4.21TVTLSFTTDD674 pKa = 3.17GSHH677 pKa = 6.86SGTPTVTLGGNSVTATNTSGNTYY700 pKa = 7.73TASYY704 pKa = 8.85TLQAGDD710 pKa = 3.5TEE712 pKa = 4.82GAVSFTIDD720 pKa = 3.36AVDD723 pKa = 3.24AAGNAMTQVTATTDD737 pKa = 3.15SSSVSFDD744 pKa = 3.03EE745 pKa = 4.17TAPTINSVSVADD757 pKa = 3.88GTYY760 pKa = 10.38KK761 pKa = 10.36IGDD764 pKa = 4.63AITITVTAGSNEE776 pKa = 4.2TGLTLAGTFNGQTLTGITDD795 pKa = 3.84NNDD798 pKa = 2.67GTYY801 pKa = 10.42SATYY805 pKa = 8.46TAVEE809 pKa = 4.29GDD811 pKa = 3.83SDD813 pKa = 4.02VADD816 pKa = 4.31AGSVTTNITLTDD828 pKa = 3.48AAGNASSATTSVTLAGEE845 pKa = 4.56SIDD848 pKa = 4.09GNSPTGSVSDD858 pKa = 4.19LVTFPTIQLEE868 pKa = 4.32ATGYY872 pKa = 5.74TTGEE876 pKa = 3.85DD877 pKa = 3.05RR878 pKa = 11.84YY879 pKa = 10.9VALEE883 pKa = 4.09SVGSDD888 pKa = 2.61GGYY891 pKa = 8.51VVAWEE896 pKa = 4.71GEE898 pKa = 4.28YY899 pKa = 11.1GSYY902 pKa = 9.47PLKK905 pKa = 10.6SIYY908 pKa = 9.67VQQFNADD915 pKa = 3.57GTTTGYY921 pKa = 10.95SPVQLDD927 pKa = 4.51GIGVTDD933 pKa = 3.76SYY935 pKa = 11.78DD936 pKa = 3.51YY937 pKa = 10.71RR938 pKa = 11.84PKK940 pKa = 9.49ITALEE945 pKa = 4.1SGGYY949 pKa = 8.53VVTWSGLDD957 pKa = 3.45DD958 pKa = 4.97SGDD961 pKa = 3.41NDD963 pKa = 3.52TSVFVQQFNADD974 pKa = 3.72GTTTGYY980 pKa = 11.28DD981 pKa = 3.6PVQLEE986 pKa = 4.28ATGYY990 pKa = 7.46TAGEE994 pKa = 4.17DD995 pKa = 3.97DD996 pKa = 4.35EE997 pKa = 4.75PQITSLGSSGEE1008 pKa = 4.04YY1009 pKa = 9.08VVIWRR1014 pKa = 11.84GQNSSTSSDD1023 pKa = 2.65RR1024 pKa = 11.84SIFVQQFNANGTVKK1038 pKa = 10.7NDD1040 pKa = 3.35TPVILEE1046 pKa = 4.08AHH1048 pKa = 6.66GSWDD1052 pKa = 3.3QYY1054 pKa = 9.09QQITTLNSGGYY1065 pKa = 8.41VVTWTGLDD1073 pKa = 3.3GTIYY1077 pKa = 10.4PDD1079 pKa = 3.4TDD1081 pKa = 3.29YY1082 pKa = 11.37SIYY1085 pKa = 10.3VQQFNADD1092 pKa = 3.59GTRR1095 pKa = 11.84TGDD1098 pKa = 3.51TPVKK1102 pKa = 10.83LEE1104 pKa = 4.01GTGNTTGGDD1113 pKa = 3.2EE1114 pKa = 4.3HH1115 pKa = 8.04SQVTALDD1122 pKa = 3.78FNGGYY1127 pKa = 8.93VVAWKK1132 pKa = 9.61GQNSDD1137 pKa = 3.14NRR1139 pKa = 11.84DD1140 pKa = 3.7SIFVQQFDD1148 pKa = 4.1ANGTITGYY1156 pKa = 9.07TPVQLNSHH1164 pKa = 6.08SNDD1167 pKa = 2.97RR1168 pKa = 11.84DD1169 pKa = 3.64VQIAAVGSSGQYY1181 pKa = 7.72VATWVGMDD1189 pKa = 3.59SNYY1192 pKa = 10.72SYY1194 pKa = 11.18DD1195 pKa = 3.48IFVQQFNASGTTDD1208 pKa = 2.73GFTTVKK1214 pKa = 10.5LEE1216 pKa = 4.54SINNPDD1222 pKa = 3.55SGNGAKK1228 pKa = 10.12PKK1230 pKa = 10.06IAALGDD1236 pKa = 3.34SGRR1239 pKa = 11.84YY1240 pKa = 8.63LVTWYY1245 pKa = 10.11DD1246 pKa = 3.4YY1247 pKa = 11.46DD1248 pKa = 3.92EE1249 pKa = 5.44EE1250 pKa = 6.28GDD1252 pKa = 3.61EE1253 pKa = 5.6SIFLQAFNADD1263 pKa = 3.45GTTAGPTQQFEE1274 pKa = 4.52ATGNTTGSDD1283 pKa = 3.45QYY1285 pKa = 11.55PEE1287 pKa = 4.03VLALGDD1293 pKa = 3.32SGAFVLAWHH1302 pKa = 6.61GRR1304 pKa = 11.84DD1305 pKa = 3.3SDD1307 pKa = 4.13DD1308 pKa = 4.51DD1309 pKa = 4.1VSIFVKK1315 pKa = 10.54QFFIDD1320 pKa = 3.65GMVRR1324 pKa = 11.84AQSSEE1329 pKa = 3.64AATAYY1334 pKa = 10.07LVKK1337 pKa = 10.27DD1338 pKa = 4.23TITVNSVTDD1347 pKa = 3.43LTGAPDD1353 pKa = 3.47SQMNTVAITSANTDD1367 pKa = 2.67TDD1369 pKa = 4.03LPLTGLEE1376 pKa = 4.03EE1377 pKa = 4.54GNYY1380 pKa = 9.07TLYY1383 pKa = 11.16LVDD1386 pKa = 3.34AAGNIGTVGTSVTVDD1401 pKa = 3.32HH1402 pKa = 6.97TPPNAPTISAVTDD1415 pKa = 3.65DD1416 pKa = 3.93QGSVTGTVADD1426 pKa = 5.16DD1427 pKa = 5.12GISDD1431 pKa = 3.93DD1432 pKa = 4.14TILTVRR1438 pKa = 11.84VSLDD1442 pKa = 3.17STNALSGDD1450 pKa = 3.82SVQLYY1455 pKa = 10.77DD1456 pKa = 4.16GFSALGTAVTLTSTDD1471 pKa = 2.73ISNGYY1476 pKa = 9.6VDD1478 pKa = 5.05IDD1480 pKa = 4.28TPTLTAGTSYY1490 pKa = 11.25NFRR1493 pKa = 11.84ANITDD1498 pKa = 3.19AFGNIGEE1505 pKa = 4.54FSTDD1509 pKa = 2.66GAADD1513 pKa = 3.56VTIDD1517 pKa = 3.27TTAPDD1522 pKa = 3.97GNVVSTTAFVTLEE1535 pKa = 3.96ASGVTNMDD1543 pKa = 3.57DD1544 pKa = 4.1RR1545 pKa = 11.84NPQISAIGSNGEE1557 pKa = 4.38YY1558 pKa = 10.25VVTWTGGDD1566 pKa = 3.7GPDD1569 pKa = 5.09DD1570 pKa = 4.79YY1571 pKa = 11.74DD1572 pKa = 4.47DD1573 pKa = 4.77SIYY1576 pKa = 10.67VQHH1579 pKa = 6.9FNTDD1583 pKa = 2.96GTITGKK1589 pKa = 10.48AQVKK1593 pKa = 10.24LEE1595 pKa = 4.26STGVTDD1601 pKa = 5.17GGDD1604 pKa = 3.38SAPQVTSVGISGEE1617 pKa = 4.49YY1618 pKa = 8.94VVTWVGNDD1626 pKa = 3.55GPDD1629 pKa = 4.22DD1630 pKa = 4.74YY1631 pKa = 11.75DD1632 pKa = 4.4DD1633 pKa = 4.77SIYY1636 pKa = 10.63VQHH1639 pKa = 6.62FNADD1643 pKa = 3.27GTITGKK1649 pKa = 10.51AQVKK1653 pKa = 10.24LEE1655 pKa = 4.24STGVTDD1661 pKa = 5.59RR1662 pKa = 11.84GDD1664 pKa = 3.37EE1665 pKa = 4.25APQVTSVGISGEE1677 pKa = 4.49YY1678 pKa = 8.94VVTWVGNDD1686 pKa = 3.46GPGDD1690 pKa = 4.06YY1691 pKa = 10.52DD1692 pKa = 4.01DD1693 pKa = 5.56SIYY1696 pKa = 10.67VQHH1699 pKa = 6.62FNADD1703 pKa = 3.27GTITGKK1709 pKa = 10.47AQVKK1713 pKa = 9.97LEE1715 pKa = 4.14PTGVTDD1721 pKa = 6.01RR1722 pKa = 11.84GDD1724 pKa = 3.57KK1725 pKa = 10.71DD1726 pKa = 3.66PQVTSVGISGEE1737 pKa = 4.49YY1738 pKa = 8.94VVTWVGNDD1746 pKa = 3.43GSDD1749 pKa = 3.04EE1750 pKa = 4.42SIYY1753 pKa = 10.16VQHH1756 pKa = 6.6FNADD1760 pKa = 3.34GTITGKK1766 pKa = 10.27DD1767 pKa = 3.47QVKK1770 pKa = 10.64LEE1772 pKa = 4.19PTGQLSGDD1780 pKa = 3.82DD1781 pKa = 4.34LRR1783 pKa = 11.84PQVSAVGSNGEE1794 pKa = 4.28YY1795 pKa = 9.75VVVWDD1800 pKa = 5.26GVDD1803 pKa = 3.24ADD1805 pKa = 3.91GDD1807 pKa = 3.54RR1808 pKa = 11.84SVFVQRR1814 pKa = 11.84FNADD1818 pKa = 2.9GTKK1821 pKa = 10.64NGTEE1825 pKa = 4.08VLLEE1829 pKa = 4.01ATGVTDD1835 pKa = 4.78QSDD1838 pKa = 3.92MQAKK1842 pKa = 9.24IAAIGSSGEE1851 pKa = 4.67FVVTWPGKK1859 pKa = 10.41DD1860 pKa = 3.11SDD1862 pKa = 4.17GDD1864 pKa = 3.62DD1865 pKa = 4.04SIFVQHH1871 pKa = 6.36FNADD1875 pKa = 3.41GSTTNHH1881 pKa = 6.79SIVQLEE1887 pKa = 4.6AIGVTDD1893 pKa = 5.72DD1894 pKa = 4.46RR1895 pKa = 11.84DD1896 pKa = 3.71EE1897 pKa = 4.56NPQITALANSGYY1909 pKa = 10.06VVTWLGDD1916 pKa = 3.46DD1917 pKa = 4.54ANNDD1921 pKa = 3.53DD1922 pKa = 5.3QVYY1925 pKa = 9.39LQQFNADD1932 pKa = 3.54GSKK1935 pKa = 10.27SGSVTLVEE1943 pKa = 4.27VGNNEE1948 pKa = 4.09IASKK1952 pKa = 10.5GYY1954 pKa = 9.81PEE1956 pKa = 4.03IAAVGIGSQFAITSRR1971 pKa = 11.84GDD1973 pKa = 3.27DD1974 pKa = 3.25SGGDD1978 pKa = 3.21DD1979 pKa = 3.23SVYY1982 pKa = 10.55VRR1984 pKa = 11.84QFDD1987 pKa = 3.52ASTVFNNSKK1996 pKa = 8.37TVQVQSDD2003 pKa = 4.75GPGMAYY2009 pKa = 10.6LIKK2012 pKa = 10.75DD2013 pKa = 3.57SVTINSAADD2022 pKa = 3.55FSGVTDD2028 pKa = 3.58SQINSVSITSANTFTDD2044 pKa = 3.58LTLSGLEE2051 pKa = 3.9AGDD2054 pKa = 3.58YY2055 pKa = 10.82KK2056 pKa = 11.2LYY2058 pKa = 11.01LVDD2061 pKa = 3.53AAGNLGITTDD2071 pKa = 4.47GLTIDD2076 pKa = 4.0NTAPTASVTAATIGSSGNAVVQSTEE2101 pKa = 3.47AGTAFLVKK2109 pKa = 10.57DD2110 pKa = 4.18SITVNSLSDD2119 pKa = 3.13ITGASDD2125 pKa = 5.1NNWNWVTISTASSDD2139 pKa = 3.95TNLAAAGLVDD2149 pKa = 4.58GDD2151 pKa = 4.11YY2152 pKa = 11.05KK2153 pKa = 11.51VYY2155 pKa = 8.47TTDD2158 pKa = 3.17SAGNLSTVSTNSVTIDD2174 pKa = 3.35TTPPTTTYY2182 pKa = 10.91SGVSYY2187 pKa = 11.51DD2188 pKa = 3.6MASNTLTLTGTGINSMLEE2206 pKa = 4.07SGEE2209 pKa = 4.91DD2210 pKa = 3.15ATTDD2214 pKa = 3.08IKK2216 pKa = 11.45DD2217 pKa = 3.48RR2218 pKa = 11.84LDD2220 pKa = 3.07WSKK2223 pKa = 11.19LVWTFNNNTSYY2234 pKa = 11.61DD2235 pKa = 3.78FGSGDD2240 pKa = 3.78LGNNTGDD2247 pKa = 5.15DD2248 pKa = 3.74ILSVKK2253 pKa = 10.3AVSDD2257 pKa = 3.49TSLEE2261 pKa = 3.93ITFTDD2266 pKa = 4.53DD2267 pKa = 3.33AANACGSGLEE2277 pKa = 4.31SFFNGFDD2284 pKa = 3.46TSNPNYY2290 pKa = 10.74APFSDD2295 pKa = 3.65GHH2297 pKa = 6.42KK2298 pKa = 9.49LTVTAGFSRR2307 pKa = 11.84DD2308 pKa = 3.64SNGNAATGDD2317 pKa = 4.03GYY2319 pKa = 11.74VNPDD2323 pKa = 3.0TSIVVFDD2330 pKa = 5.32LIQGVSSSHH2339 pKa = 6.25SSRR2342 pKa = 11.84TFDD2345 pKa = 3.97ANTSYY2350 pKa = 10.76TIYY2353 pKa = 10.62IRR2355 pKa = 11.84MDD2357 pKa = 3.5SDD2359 pKa = 3.71SSTLSTAGSGPGTWGTWSSANNLGTDD2385 pKa = 3.62DD2386 pKa = 3.89KK2387 pKa = 11.64VIFVGSGSVIKK2398 pKa = 10.9NNSNPAQEE2406 pKa = 3.85VTFYY2410 pKa = 10.84YY2411 pKa = 10.55KK2412 pKa = 10.44SAGLLEE2418 pKa = 4.91LNYY2421 pKa = 10.39SLNSRR2426 pKa = 11.84TVQLYY2431 pKa = 9.05IYY2433 pKa = 9.7LGVGSIARR2441 pKa = 11.84NNYY2444 pKa = 9.63ASSEE2448 pKa = 4.17KK2449 pKa = 10.51LWDD2452 pKa = 3.77GNISVGDD2459 pKa = 4.03LRR2461 pKa = 11.84PTTYY2465 pKa = 9.72STGKK2469 pKa = 10.47FNDD2472 pKa = 3.93FVLTTMPNGILTSQGLVV2489 pKa = 3.12
Molecular weight: 255.77 kDa
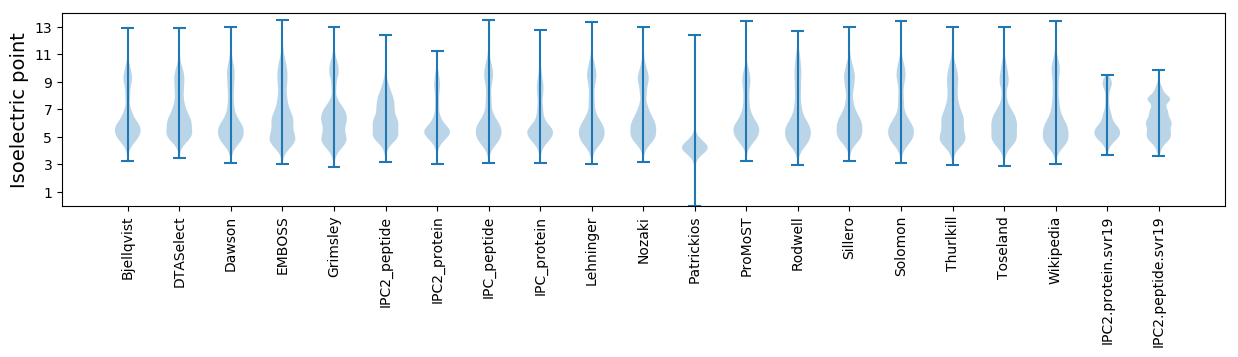
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C2HI14|A0A5C2HI14_9GAMM GTP diphosphokinase OS=Ectothiorhodospiraceae bacterium BW-2 OX=947515 GN=D5085_05745 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNIKK11 pKa = 10.05RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 6.03GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 10.06SVGGRR28 pKa = 11.84KK29 pKa = 8.25VLKK32 pKa = 10.25ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.31GRR39 pKa = 11.84ARR41 pKa = 11.84LALL44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNIKK11 pKa = 10.05RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 6.03GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 10.06SVGGRR28 pKa = 11.84KK29 pKa = 8.25VLKK32 pKa = 10.25ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.31GRR39 pKa = 11.84ARR41 pKa = 11.84LALL44 pKa = 3.93
Molecular weight: 5.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1182593 |
33 |
6638 |
343.3 |
38.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.061 ± 0.052 | 0.958 ± 0.016 |
5.127 ± 0.037 | 6.697 ± 0.045 |
3.474 ± 0.028 | 6.752 ± 0.052 |
2.495 ± 0.032 | 5.848 ± 0.033 |
3.549 ± 0.045 | 11.693 ± 0.076 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.28 ± 0.024 | 3.408 ± 0.04 |
4.472 ± 0.03 | 5.81 ± 0.06 |
6.274 ± 0.062 | 6.321 ± 0.052 |
5.335 ± 0.082 | 6.107 ± 0.039 |
1.349 ± 0.018 | 2.993 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
