
Hendra virus (isolate Horse/Autralia/Hendra/1994)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Henipavirus; Hendra henipavirus
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
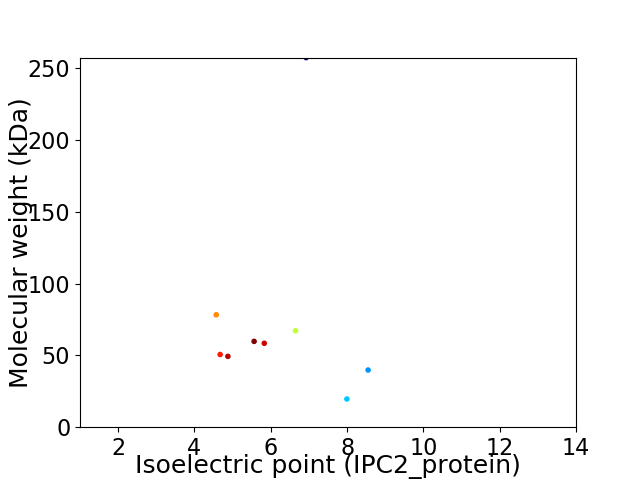
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
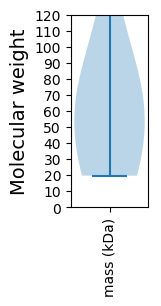
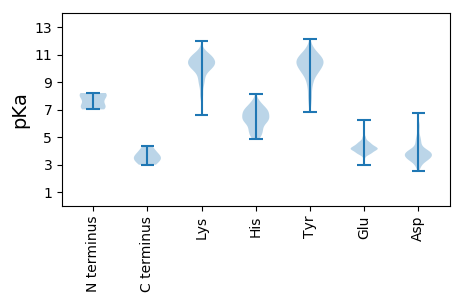
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|O55779|C_HENDH Protein C OS=Hendra virus (isolate Horse/Autralia/Hendra/1994) OX=928303 GN=P/V/C PE=2 SV=1
MM1 pKa = 8.2DD2 pKa = 5.84KK3 pKa = 11.15LDD5 pKa = 4.35LVNDD9 pKa = 4.02GLDD12 pKa = 3.67IIDD15 pKa = 6.0FIQKK19 pKa = 7.96NQKK22 pKa = 9.6EE23 pKa = 4.2IQKK26 pKa = 9.07TYY28 pKa = 10.35GRR30 pKa = 11.84SSIQQPSTKK39 pKa = 10.23DD40 pKa = 2.92RR41 pKa = 11.84TRR43 pKa = 11.84AWEE46 pKa = 4.59DD47 pKa = 3.33FLQSTSGEE55 pKa = 4.16HH56 pKa = 5.34EE57 pKa = 4.04QAEE60 pKa = 4.25GGMPKK65 pKa = 10.18NDD67 pKa = 3.35GGTEE71 pKa = 3.91GRR73 pKa = 11.84NVEE76 pKa = 4.49DD77 pKa = 4.08LSSVTSSDD85 pKa = 2.88GTIGQRR91 pKa = 11.84VSNTRR96 pKa = 11.84AWAEE100 pKa = 4.15DD101 pKa = 3.71PDD103 pKa = 5.85DD104 pKa = 3.99IQLDD108 pKa = 3.79PMVTDD113 pKa = 3.77VVYY116 pKa = 10.38HH117 pKa = 5.5DD118 pKa = 5.08HH119 pKa = 7.03GGEE122 pKa = 4.34CTGHH126 pKa = 6.32GPSSSPEE133 pKa = 4.11RR134 pKa = 11.84GWSYY138 pKa = 11.35HH139 pKa = 4.99MSGTHH144 pKa = 6.83DD145 pKa = 3.6GNVRR149 pKa = 11.84AVPDD153 pKa = 3.84TKK155 pKa = 10.96VLPNAPKK162 pKa = 7.97TTVPEE167 pKa = 4.04EE168 pKa = 3.8VRR170 pKa = 11.84EE171 pKa = 3.74IDD173 pKa = 5.55LIGLEE178 pKa = 4.8DD179 pKa = 3.68KK180 pKa = 10.69FASAGLNPAAVPFVPKK196 pKa = 10.41NQSTPTEE203 pKa = 4.22EE204 pKa = 4.17PPVIPEE210 pKa = 3.8YY211 pKa = 10.91YY212 pKa = 9.21YY213 pKa = 10.92GSGRR217 pKa = 11.84RR218 pKa = 11.84GDD220 pKa = 3.94LSKK223 pKa = 11.06SPPRR227 pKa = 11.84GNVNLDD233 pKa = 3.39SIKK236 pKa = 10.28IYY238 pKa = 10.63TSDD241 pKa = 5.66DD242 pKa = 3.27EE243 pKa = 6.1DD244 pKa = 4.12EE245 pKa = 4.57NQLEE249 pKa = 4.37YY250 pKa = 10.84EE251 pKa = 4.37DD252 pKa = 5.24EE253 pKa = 4.3FAKK256 pKa = 10.76SSSEE260 pKa = 4.06VVIDD264 pKa = 3.71TTPEE268 pKa = 4.12DD269 pKa = 3.52NDD271 pKa = 4.54SINQEE276 pKa = 4.17EE277 pKa = 4.98VVGDD281 pKa = 3.99PSDD284 pKa = 3.27QGLEE288 pKa = 4.08HH289 pKa = 7.35PFPLGKK295 pKa = 10.19FPEE298 pKa = 4.49KK299 pKa = 10.9EE300 pKa = 3.77EE301 pKa = 4.29TPDD304 pKa = 3.46VRR306 pKa = 11.84RR307 pKa = 11.84KK308 pKa = 10.48DD309 pKa = 3.98SLMQDD314 pKa = 2.74SCKK317 pKa = 10.41RR318 pKa = 11.84GGVPKK323 pKa = 10.14RR324 pKa = 11.84LPMLSEE330 pKa = 4.08EE331 pKa = 4.65FEE333 pKa = 4.69CSGSDD338 pKa = 3.89DD339 pKa = 4.82PIIQEE344 pKa = 4.03LEE346 pKa = 4.14RR347 pKa = 11.84EE348 pKa = 4.45GSHH351 pKa = 6.9PGGSLRR357 pKa = 11.84LRR359 pKa = 11.84EE360 pKa = 4.37PPQSSGNSRR369 pKa = 11.84NQPDD373 pKa = 3.87RR374 pKa = 11.84QLKK377 pKa = 8.89TGDD380 pKa = 3.34AASPGGVQRR389 pKa = 11.84PGTPMPKK396 pKa = 9.92SRR398 pKa = 11.84IMPIKK403 pKa = 10.11KK404 pKa = 8.71GTDD407 pKa = 2.84AKK409 pKa = 10.02SQYY412 pKa = 10.86VGTEE416 pKa = 3.95DD417 pKa = 3.92VPGSKK422 pKa = 9.95SGATRR427 pKa = 11.84YY428 pKa = 9.64VRR430 pKa = 11.84GLPPNQEE437 pKa = 4.37SKK439 pKa = 11.0SVTAEE444 pKa = 3.86NVQLSAPSAVTRR456 pKa = 11.84NEE458 pKa = 3.7GHH460 pKa = 6.13DD461 pKa = 3.58QEE463 pKa = 4.66VTSNEE468 pKa = 3.82DD469 pKa = 3.47SLDD472 pKa = 3.55DD473 pKa = 4.32KK474 pKa = 11.7YY475 pKa = 11.71IMPSDD480 pKa = 4.12DD481 pKa = 3.54FANTFLPHH489 pKa = 6.27DD490 pKa = 3.98TDD492 pKa = 3.45RR493 pKa = 11.84LNYY496 pKa = 9.78HH497 pKa = 7.13ADD499 pKa = 3.27HH500 pKa = 6.8LNDD503 pKa = 3.59YY504 pKa = 10.8DD505 pKa = 6.15LEE507 pKa = 4.53TLCEE511 pKa = 4.02EE512 pKa = 4.32SVLMGIVNAIKK523 pKa = 10.6LINIDD528 pKa = 2.99MRR530 pKa = 11.84LNHH533 pKa = 6.89IEE535 pKa = 4.08EE536 pKa = 4.13QMKK539 pKa = 9.97EE540 pKa = 3.54IPKK543 pKa = 9.95IINKK547 pKa = 9.12IDD549 pKa = 3.73SIDD552 pKa = 3.62RR553 pKa = 11.84VLAKK557 pKa = 10.14TNTALSTIEE566 pKa = 3.82GHH568 pKa = 6.27LVSMMIMIPGKK579 pKa = 10.71GKK581 pKa = 10.41GEE583 pKa = 4.01RR584 pKa = 11.84KK585 pKa = 10.06GKK587 pKa = 8.56TNPEE591 pKa = 4.17LKK593 pKa = 10.27PVIGRR598 pKa = 11.84NILEE602 pKa = 4.11QQEE605 pKa = 4.59LFSFDD610 pKa = 3.12NLKK613 pKa = 10.5NFRR616 pKa = 11.84DD617 pKa = 4.0GSLTDD622 pKa = 3.4EE623 pKa = 5.05PYY625 pKa = 11.12GGVARR630 pKa = 11.84IRR632 pKa = 11.84DD633 pKa = 3.99DD634 pKa = 5.43LILPEE639 pKa = 4.98LNFSEE644 pKa = 4.7TNASQFVPLADD655 pKa = 4.39DD656 pKa = 3.73ASKK659 pKa = 11.32DD660 pKa = 3.65VVRR663 pKa = 11.84TMIRR667 pKa = 11.84THH669 pKa = 5.8IKK671 pKa = 10.32DD672 pKa = 3.27RR673 pKa = 11.84EE674 pKa = 4.18LRR676 pKa = 11.84SEE678 pKa = 4.36LMDD681 pKa = 3.62YY682 pKa = 11.1LNRR685 pKa = 11.84AEE687 pKa = 4.16TDD689 pKa = 3.46EE690 pKa = 4.25EE691 pKa = 4.33VQEE694 pKa = 4.36VANTVNDD701 pKa = 4.37IIDD704 pKa = 3.68GNII707 pKa = 2.98
MM1 pKa = 8.2DD2 pKa = 5.84KK3 pKa = 11.15LDD5 pKa = 4.35LVNDD9 pKa = 4.02GLDD12 pKa = 3.67IIDD15 pKa = 6.0FIQKK19 pKa = 7.96NQKK22 pKa = 9.6EE23 pKa = 4.2IQKK26 pKa = 9.07TYY28 pKa = 10.35GRR30 pKa = 11.84SSIQQPSTKK39 pKa = 10.23DD40 pKa = 2.92RR41 pKa = 11.84TRR43 pKa = 11.84AWEE46 pKa = 4.59DD47 pKa = 3.33FLQSTSGEE55 pKa = 4.16HH56 pKa = 5.34EE57 pKa = 4.04QAEE60 pKa = 4.25GGMPKK65 pKa = 10.18NDD67 pKa = 3.35GGTEE71 pKa = 3.91GRR73 pKa = 11.84NVEE76 pKa = 4.49DD77 pKa = 4.08LSSVTSSDD85 pKa = 2.88GTIGQRR91 pKa = 11.84VSNTRR96 pKa = 11.84AWAEE100 pKa = 4.15DD101 pKa = 3.71PDD103 pKa = 5.85DD104 pKa = 3.99IQLDD108 pKa = 3.79PMVTDD113 pKa = 3.77VVYY116 pKa = 10.38HH117 pKa = 5.5DD118 pKa = 5.08HH119 pKa = 7.03GGEE122 pKa = 4.34CTGHH126 pKa = 6.32GPSSSPEE133 pKa = 4.11RR134 pKa = 11.84GWSYY138 pKa = 11.35HH139 pKa = 4.99MSGTHH144 pKa = 6.83DD145 pKa = 3.6GNVRR149 pKa = 11.84AVPDD153 pKa = 3.84TKK155 pKa = 10.96VLPNAPKK162 pKa = 7.97TTVPEE167 pKa = 4.04EE168 pKa = 3.8VRR170 pKa = 11.84EE171 pKa = 3.74IDD173 pKa = 5.55LIGLEE178 pKa = 4.8DD179 pKa = 3.68KK180 pKa = 10.69FASAGLNPAAVPFVPKK196 pKa = 10.41NQSTPTEE203 pKa = 4.22EE204 pKa = 4.17PPVIPEE210 pKa = 3.8YY211 pKa = 10.91YY212 pKa = 9.21YY213 pKa = 10.92GSGRR217 pKa = 11.84RR218 pKa = 11.84GDD220 pKa = 3.94LSKK223 pKa = 11.06SPPRR227 pKa = 11.84GNVNLDD233 pKa = 3.39SIKK236 pKa = 10.28IYY238 pKa = 10.63TSDD241 pKa = 5.66DD242 pKa = 3.27EE243 pKa = 6.1DD244 pKa = 4.12EE245 pKa = 4.57NQLEE249 pKa = 4.37YY250 pKa = 10.84EE251 pKa = 4.37DD252 pKa = 5.24EE253 pKa = 4.3FAKK256 pKa = 10.76SSSEE260 pKa = 4.06VVIDD264 pKa = 3.71TTPEE268 pKa = 4.12DD269 pKa = 3.52NDD271 pKa = 4.54SINQEE276 pKa = 4.17EE277 pKa = 4.98VVGDD281 pKa = 3.99PSDD284 pKa = 3.27QGLEE288 pKa = 4.08HH289 pKa = 7.35PFPLGKK295 pKa = 10.19FPEE298 pKa = 4.49KK299 pKa = 10.9EE300 pKa = 3.77EE301 pKa = 4.29TPDD304 pKa = 3.46VRR306 pKa = 11.84RR307 pKa = 11.84KK308 pKa = 10.48DD309 pKa = 3.98SLMQDD314 pKa = 2.74SCKK317 pKa = 10.41RR318 pKa = 11.84GGVPKK323 pKa = 10.14RR324 pKa = 11.84LPMLSEE330 pKa = 4.08EE331 pKa = 4.65FEE333 pKa = 4.69CSGSDD338 pKa = 3.89DD339 pKa = 4.82PIIQEE344 pKa = 4.03LEE346 pKa = 4.14RR347 pKa = 11.84EE348 pKa = 4.45GSHH351 pKa = 6.9PGGSLRR357 pKa = 11.84LRR359 pKa = 11.84EE360 pKa = 4.37PPQSSGNSRR369 pKa = 11.84NQPDD373 pKa = 3.87RR374 pKa = 11.84QLKK377 pKa = 8.89TGDD380 pKa = 3.34AASPGGVQRR389 pKa = 11.84PGTPMPKK396 pKa = 9.92SRR398 pKa = 11.84IMPIKK403 pKa = 10.11KK404 pKa = 8.71GTDD407 pKa = 2.84AKK409 pKa = 10.02SQYY412 pKa = 10.86VGTEE416 pKa = 3.95DD417 pKa = 3.92VPGSKK422 pKa = 9.95SGATRR427 pKa = 11.84YY428 pKa = 9.64VRR430 pKa = 11.84GLPPNQEE437 pKa = 4.37SKK439 pKa = 11.0SVTAEE444 pKa = 3.86NVQLSAPSAVTRR456 pKa = 11.84NEE458 pKa = 3.7GHH460 pKa = 6.13DD461 pKa = 3.58QEE463 pKa = 4.66VTSNEE468 pKa = 3.82DD469 pKa = 3.47SLDD472 pKa = 3.55DD473 pKa = 4.32KK474 pKa = 11.7YY475 pKa = 11.71IMPSDD480 pKa = 4.12DD481 pKa = 3.54FANTFLPHH489 pKa = 6.27DD490 pKa = 3.98TDD492 pKa = 3.45RR493 pKa = 11.84LNYY496 pKa = 9.78HH497 pKa = 7.13ADD499 pKa = 3.27HH500 pKa = 6.8LNDD503 pKa = 3.59YY504 pKa = 10.8DD505 pKa = 6.15LEE507 pKa = 4.53TLCEE511 pKa = 4.02EE512 pKa = 4.32SVLMGIVNAIKK523 pKa = 10.6LINIDD528 pKa = 2.99MRR530 pKa = 11.84LNHH533 pKa = 6.89IEE535 pKa = 4.08EE536 pKa = 4.13QMKK539 pKa = 9.97EE540 pKa = 3.54IPKK543 pKa = 9.95IINKK547 pKa = 9.12IDD549 pKa = 3.73SIDD552 pKa = 3.62RR553 pKa = 11.84VLAKK557 pKa = 10.14TNTALSTIEE566 pKa = 3.82GHH568 pKa = 6.27LVSMMIMIPGKK579 pKa = 10.71GKK581 pKa = 10.41GEE583 pKa = 4.01RR584 pKa = 11.84KK585 pKa = 10.06GKK587 pKa = 8.56TNPEE591 pKa = 4.17LKK593 pKa = 10.27PVIGRR598 pKa = 11.84NILEE602 pKa = 4.11QQEE605 pKa = 4.59LFSFDD610 pKa = 3.12NLKK613 pKa = 10.5NFRR616 pKa = 11.84DD617 pKa = 4.0GSLTDD622 pKa = 3.4EE623 pKa = 5.05PYY625 pKa = 11.12GGVARR630 pKa = 11.84IRR632 pKa = 11.84DD633 pKa = 3.99DD634 pKa = 5.43LILPEE639 pKa = 4.98LNFSEE644 pKa = 4.7TNASQFVPLADD655 pKa = 4.39DD656 pKa = 3.73ASKK659 pKa = 11.32DD660 pKa = 3.65VVRR663 pKa = 11.84TMIRR667 pKa = 11.84THH669 pKa = 5.8IKK671 pKa = 10.32DD672 pKa = 3.27RR673 pKa = 11.84EE674 pKa = 4.18LRR676 pKa = 11.84SEE678 pKa = 4.36LMDD681 pKa = 3.62YY682 pKa = 11.1LNRR685 pKa = 11.84AEE687 pKa = 4.16TDD689 pKa = 3.46EE690 pKa = 4.25EE691 pKa = 4.33VQEE694 pKa = 4.36VANTVNDD701 pKa = 4.37IIDD704 pKa = 3.68GNII707 pKa = 2.98
Molecular weight: 78.31 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|O89342|FUS_HENDH Fusion glycoprotein F0 OS=Hendra virus (isolate Horse/Autralia/Hendra/1994) OX=928303 GN=F PE=1 SV=2
MM1 pKa = 7.83DD2 pKa = 5.04FSVSDD7 pKa = 3.91NLDD10 pKa = 3.55DD11 pKa = 5.46PIEE14 pKa = 4.41GVSDD18 pKa = 4.4FSPTSWEE25 pKa = 3.72NGGYY29 pKa = 9.56LDD31 pKa = 4.74KK32 pKa = 11.47VEE34 pKa = 4.56PEE36 pKa = 3.8IDD38 pKa = 3.15KK39 pKa = 10.67HH40 pKa = 8.13GSMIPKK46 pKa = 9.78YY47 pKa = 10.43KK48 pKa = 10.14IYY50 pKa = 10.47TPGANEE56 pKa = 3.92RR57 pKa = 11.84KK58 pKa = 9.29FNNYY62 pKa = 8.05MYY64 pKa = 8.14MICYY68 pKa = 9.91GFVEE72 pKa = 4.43DD73 pKa = 4.87VEE75 pKa = 4.66RR76 pKa = 11.84SPEE79 pKa = 3.72SGKK82 pKa = 10.18RR83 pKa = 11.84KK84 pKa = 9.73KK85 pKa = 10.13IRR87 pKa = 11.84TIAAYY92 pKa = 8.88PLGVGKK98 pKa = 8.55STSHH102 pKa = 6.1PQDD105 pKa = 3.82LLEE108 pKa = 4.51EE109 pKa = 4.4LCSLKK114 pKa = 9.7VTVRR118 pKa = 11.84RR119 pKa = 11.84TAGATEE125 pKa = 4.39KK126 pKa = 10.48IVFGSSGPLHH136 pKa = 6.95HH137 pKa = 7.19LLPWKK142 pKa = 10.39KK143 pKa = 9.78ILTGGSIFNAVKK155 pKa = 9.82VCRR158 pKa = 11.84NVDD161 pKa = 3.61QIQLEE166 pKa = 4.4NQQSLRR172 pKa = 11.84IFFLSITKK180 pKa = 10.61LNDD183 pKa = 2.92SGIYY187 pKa = 8.39MIPRR191 pKa = 11.84TMLEE195 pKa = 3.67FRR197 pKa = 11.84RR198 pKa = 11.84NNAIAFNLLVYY209 pKa = 10.11LKK211 pKa = 10.15IDD213 pKa = 3.72ADD215 pKa = 3.97LAKK218 pKa = 10.71AGIQGSFDD226 pKa = 3.58KK227 pKa = 11.15DD228 pKa = 3.39GTKK231 pKa = 10.06VASFMLHH238 pKa = 6.3LGNFVRR244 pKa = 11.84RR245 pKa = 11.84AGKK248 pKa = 9.46YY249 pKa = 9.93YY250 pKa = 10.36SVEE253 pKa = 3.81YY254 pKa = 10.06CKK256 pKa = 10.67RR257 pKa = 11.84KK258 pKa = 8.62IDD260 pKa = 3.75RR261 pKa = 11.84MKK263 pKa = 10.86LQFSLGSIGGLSLHH277 pKa = 6.47IKK279 pKa = 9.92INGVISKK286 pKa = 10.48RR287 pKa = 11.84LFAQMGFQKK296 pKa = 10.44NLCFSLMDD304 pKa = 3.9INPWLNRR311 pKa = 11.84LTWNNSCEE319 pKa = 4.12ISRR322 pKa = 11.84VAAVLQPSVPRR333 pKa = 11.84EE334 pKa = 3.42FMIYY338 pKa = 10.68DD339 pKa = 3.94DD340 pKa = 4.73VFIDD344 pKa = 3.65NTGKK348 pKa = 10.1ILKK351 pKa = 9.78GG352 pKa = 3.34
MM1 pKa = 7.83DD2 pKa = 5.04FSVSDD7 pKa = 3.91NLDD10 pKa = 3.55DD11 pKa = 5.46PIEE14 pKa = 4.41GVSDD18 pKa = 4.4FSPTSWEE25 pKa = 3.72NGGYY29 pKa = 9.56LDD31 pKa = 4.74KK32 pKa = 11.47VEE34 pKa = 4.56PEE36 pKa = 3.8IDD38 pKa = 3.15KK39 pKa = 10.67HH40 pKa = 8.13GSMIPKK46 pKa = 9.78YY47 pKa = 10.43KK48 pKa = 10.14IYY50 pKa = 10.47TPGANEE56 pKa = 3.92RR57 pKa = 11.84KK58 pKa = 9.29FNNYY62 pKa = 8.05MYY64 pKa = 8.14MICYY68 pKa = 9.91GFVEE72 pKa = 4.43DD73 pKa = 4.87VEE75 pKa = 4.66RR76 pKa = 11.84SPEE79 pKa = 3.72SGKK82 pKa = 10.18RR83 pKa = 11.84KK84 pKa = 9.73KK85 pKa = 10.13IRR87 pKa = 11.84TIAAYY92 pKa = 8.88PLGVGKK98 pKa = 8.55STSHH102 pKa = 6.1PQDD105 pKa = 3.82LLEE108 pKa = 4.51EE109 pKa = 4.4LCSLKK114 pKa = 9.7VTVRR118 pKa = 11.84RR119 pKa = 11.84TAGATEE125 pKa = 4.39KK126 pKa = 10.48IVFGSSGPLHH136 pKa = 6.95HH137 pKa = 7.19LLPWKK142 pKa = 10.39KK143 pKa = 9.78ILTGGSIFNAVKK155 pKa = 9.82VCRR158 pKa = 11.84NVDD161 pKa = 3.61QIQLEE166 pKa = 4.4NQQSLRR172 pKa = 11.84IFFLSITKK180 pKa = 10.61LNDD183 pKa = 2.92SGIYY187 pKa = 8.39MIPRR191 pKa = 11.84TMLEE195 pKa = 3.67FRR197 pKa = 11.84RR198 pKa = 11.84NNAIAFNLLVYY209 pKa = 10.11LKK211 pKa = 10.15IDD213 pKa = 3.72ADD215 pKa = 3.97LAKK218 pKa = 10.71AGIQGSFDD226 pKa = 3.58KK227 pKa = 11.15DD228 pKa = 3.39GTKK231 pKa = 10.06VASFMLHH238 pKa = 6.3LGNFVRR244 pKa = 11.84RR245 pKa = 11.84AGKK248 pKa = 9.46YY249 pKa = 9.93YY250 pKa = 10.36SVEE253 pKa = 3.81YY254 pKa = 10.06CKK256 pKa = 10.67RR257 pKa = 11.84KK258 pKa = 8.62IDD260 pKa = 3.75RR261 pKa = 11.84MKK263 pKa = 10.86LQFSLGSIGGLSLHH277 pKa = 6.47IKK279 pKa = 9.92INGVISKK286 pKa = 10.48RR287 pKa = 11.84LFAQMGFQKK296 pKa = 10.44NLCFSLMDD304 pKa = 3.9INPWLNRR311 pKa = 11.84LTWNNSCEE319 pKa = 4.12ISRR322 pKa = 11.84VAAVLQPSVPRR333 pKa = 11.84EE334 pKa = 3.42FMIYY338 pKa = 10.68DD339 pKa = 3.94DD340 pKa = 4.73VFIDD344 pKa = 3.65NTGKK348 pKa = 10.1ILKK351 pKa = 9.78GG352 pKa = 3.34
Molecular weight: 39.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6056 |
166 |
2244 |
672.9 |
75.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.64 ± 0.583 | 1.717 ± 0.299 |
6.555 ± 0.603 | 6.489 ± 0.642 |
3.104 ± 0.444 | 6.258 ± 0.764 |
1.882 ± 0.307 | 7.48 ± 0.71 |
6.093 ± 0.337 | 9.098 ± 0.852 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.725 ± 0.268 | 5.234 ± 0.267 |
5.168 ± 0.774 | 3.913 ± 0.323 |
5.433 ± 0.406 | 8.438 ± 0.266 |
5.664 ± 0.269 | 5.862 ± 0.353 |
0.958 ± 0.146 | 3.286 ± 0.362 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
