
Acidiluteibacter ferrifornacis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Cryomorphaceae; Acidiluteibacter
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
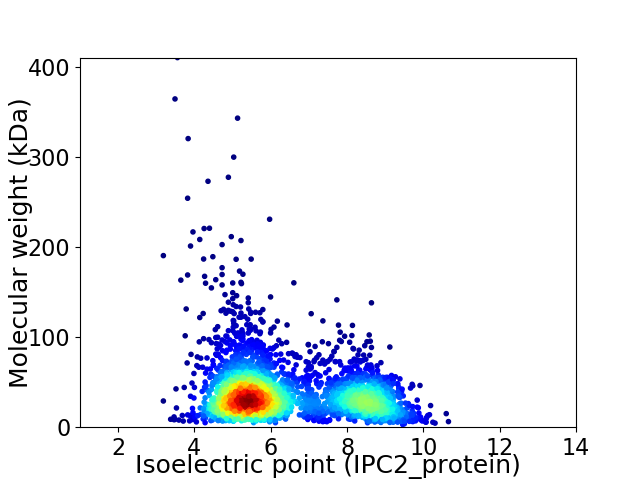
Virtual 2D-PAGE plot for 3010 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6N9NN41|A0A6N9NN41_9FLAO Methyltransferase domain-containing protein OS=Acidiluteibacter ferrifornacis OX=2692424 GN=GQN54_10940 PE=4 SV=1
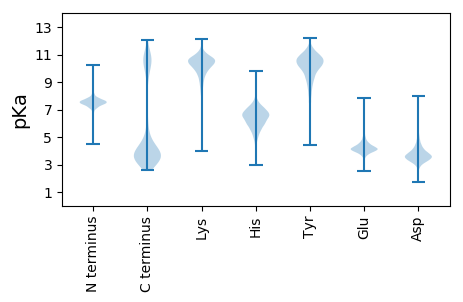
MM1 pKa = 7.82KK2 pKa = 10.32KK3 pKa = 10.06RR4 pKa = 11.84LLSVGLLLLSFAGYY18 pKa = 9.06SQIFLDD24 pKa = 4.12EE25 pKa = 4.57DD26 pKa = 4.12FEE28 pKa = 4.88TFTVGAAGPTFANGWTNSGTTTPRR52 pKa = 11.84WEE54 pKa = 4.15VEE56 pKa = 4.09DD57 pKa = 3.4ATGANEE63 pKa = 3.86NSLATGPFFDD73 pKa = 3.94ATSPTVAGGKK83 pKa = 9.28YY84 pKa = 10.19LYY86 pKa = 10.83LEE88 pKa = 4.39TSGGALNDD96 pKa = 3.55QNTLSSPPVSIPMSATGVKK115 pKa = 10.23LDD117 pKa = 3.91FAYY120 pKa = 10.78HH121 pKa = 5.5MFGASMGEE129 pKa = 4.08LYY131 pKa = 10.97VVIDD135 pKa = 3.97TNNVSDD141 pKa = 4.16TVVTLIGQQQTSGADD156 pKa = 3.42PFITSSTILTGYY168 pKa = 9.59AGKK171 pKa = 8.28TLTVKK176 pKa = 10.68FIGKK180 pKa = 9.85RR181 pKa = 11.84GTSYY185 pKa = 10.5TGDD188 pKa = 3.09ISIDD192 pKa = 3.67EE193 pKa = 4.5VKK195 pKa = 10.64LFQPVPNEE203 pKa = 3.74MGITEE208 pKa = 4.22ITTPTSSCGLTASEE222 pKa = 4.48VVTIEE227 pKa = 3.82VQNFGTSNITSFDD240 pKa = 3.15ASFSINGGTPVTEE253 pKa = 4.34TVTTTITPNSIYY265 pKa = 10.33SYY267 pKa = 10.23TFTATANLSAFGPYY281 pKa = 8.35EE282 pKa = 3.93VKK284 pKa = 10.71SYY286 pKa = 9.3VTLLNDD292 pKa = 3.59PTSSNDD298 pKa = 3.56TLVKK302 pKa = 10.67NITNIPILNGFPYY315 pKa = 10.98LEE317 pKa = 4.35TFEE320 pKa = 5.02SGPGGWISGGTNSSWALGTPAGTVINSAAGGTNSWVTNLTGTYY363 pKa = 9.72NATEE367 pKa = 3.81ASYY370 pKa = 11.27VVSPCFDD377 pKa = 3.53FTSLVAPVFSADD389 pKa = 3.31VWWNSEE395 pKa = 3.92FSWDD399 pKa = 3.51GAILQTSIDD408 pKa = 5.21GGTTWQKK415 pKa = 10.93VGANGDD421 pKa = 3.78PNNWYY426 pKa = 8.91TDD428 pKa = 3.48NTISGLTPLAPSQEE442 pKa = 4.01GWTGRR447 pKa = 11.84NSTSNGSGGWVKK459 pKa = 10.63VKK461 pKa = 10.94APLTGLGSVSGVILRR476 pKa = 11.84IAFGSDD482 pKa = 2.8GSVMDD487 pKa = 4.46EE488 pKa = 4.63GFAFDD493 pKa = 5.37NIQIYY498 pKa = 7.61DD499 pKa = 3.89TPAQDD504 pKa = 5.29AEE506 pKa = 4.5LLSITSPINGCGLTSTEE523 pKa = 3.88QAEE526 pKa = 4.41VQIVNAGSASISNFPVSITVNGGTPLTEE554 pKa = 4.07TVTATILPLDD564 pKa = 3.57TLFYY568 pKa = 10.48TFTGTVNLSTTGNYY582 pKa = 9.82SIKK585 pKa = 10.36AYY587 pKa = 9.67TGLTGDD593 pKa = 4.51GLSSNDD599 pKa = 3.2TATKK603 pKa = 10.25VVTHH607 pKa = 6.6IPIISTFPYY616 pKa = 8.68STSFEE621 pKa = 4.14NSNEE625 pKa = 3.69GWLSSGINNSWSLGVPAGSIIDD647 pKa = 3.76TASNGTKK654 pKa = 10.2AWVTNLNGVYY664 pKa = 10.59NNNEE668 pKa = 3.57QSFVVSPCLDD678 pKa = 3.58FTNLTSPLISLDD690 pKa = 3.15IQYY693 pKa = 10.99EE694 pKa = 4.15LDD696 pKa = 3.48NSNDD700 pKa = 3.32GVVLQSSIDD709 pKa = 4.49GGATWQKK716 pKa = 10.43VGAFGSPNNWYY727 pKa = 8.87TDD729 pKa = 3.37NTIVGLSSIEE739 pKa = 3.95TSQEE743 pKa = 3.12GWTGSGTSGSNGWKK757 pKa = 9.51SARR760 pKa = 11.84AALSGLGGEE769 pKa = 4.38PSVILRR775 pKa = 11.84IALGTNGFTTFEE787 pKa = 4.07GFAFDD792 pKa = 4.18NVTIQEE798 pKa = 4.43APSTDD803 pKa = 3.16AGVVDD808 pKa = 5.68IIDD811 pKa = 4.08PASGCGLSSAEE822 pKa = 4.12VVTVRR827 pKa = 11.84IVNDD831 pKa = 3.62GATNISNFPVSVVLNNGTPLTEE853 pKa = 4.2TVTATMLPNDD863 pKa = 3.71TLTYY867 pKa = 10.32TFTGTLNMSALGTYY881 pKa = 10.1SVVSYY886 pKa = 10.53TSLPLDD892 pKa = 3.59GFNVNDD898 pKa = 4.0TASKK902 pKa = 8.75TVVNIPVISGFPYY915 pKa = 9.56TEE917 pKa = 4.08SFEE920 pKa = 4.28TSAGGWTVDD929 pKa = 3.24GGTSSFALGAPIGSLINSASNGTQAWVTNLSGNYY963 pKa = 9.49LADD966 pKa = 3.23EE967 pKa = 4.58AGYY970 pKa = 10.77VKK972 pKa = 10.55SPCLDD977 pKa = 3.8FSTLIAPIISMDD989 pKa = 3.44VWWNSEE995 pKa = 3.95FSWDD999 pKa = 3.6GAVLQSSIDD1008 pKa = 4.72GGTTWQKK1015 pKa = 10.65VGSFGDD1021 pKa = 3.58PNNWYY1026 pKa = 10.18NDD1028 pKa = 3.16NSINGLNTGSPILEE1042 pKa = 4.11PSGEE1046 pKa = 3.97GWTGRR1051 pKa = 11.84NSSSNGSNGWVNATHH1066 pKa = 6.51QLTGLGGQPGVIFRR1080 pKa = 11.84VAFGSDD1086 pKa = 3.14GSVMDD1091 pKa = 4.46EE1092 pKa = 4.54GFAFDD1097 pKa = 3.9NVQIYY1102 pKa = 7.96EE1103 pKa = 4.31SPAVDD1108 pKa = 3.61IKK1110 pKa = 11.43AMEE1113 pKa = 4.05VSRR1116 pKa = 11.84PLVGCGLSATEE1127 pKa = 4.48TIAARR1132 pKa = 11.84YY1133 pKa = 7.53EE1134 pKa = 4.02NLGTDD1139 pKa = 3.42TLTNFPVAYY1148 pKa = 9.39SVNGVAITPEE1158 pKa = 4.3TFTDD1162 pKa = 3.59TLFPGDD1168 pKa = 3.45KK1169 pKa = 9.91KK1170 pKa = 11.09SYY1172 pKa = 10.65EE1173 pKa = 4.14FTTTANLSSTIQYY1186 pKa = 9.18TIEE1189 pKa = 3.93VWSAAAGDD1197 pKa = 4.9AITSNDD1203 pKa = 3.54TASVTFTNIAASTNPYY1219 pKa = 9.13PQTFDD1224 pKa = 3.48VLTDD1228 pKa = 3.71GATDD1232 pKa = 3.85FSPINWTPINAGAFDD1247 pKa = 3.72WRR1249 pKa = 11.84AEE1251 pKa = 4.15TT1252 pKa = 4.54
MM1 pKa = 7.82KK2 pKa = 10.32KK3 pKa = 10.06RR4 pKa = 11.84LLSVGLLLLSFAGYY18 pKa = 9.06SQIFLDD24 pKa = 4.12EE25 pKa = 4.57DD26 pKa = 4.12FEE28 pKa = 4.88TFTVGAAGPTFANGWTNSGTTTPRR52 pKa = 11.84WEE54 pKa = 4.15VEE56 pKa = 4.09DD57 pKa = 3.4ATGANEE63 pKa = 3.86NSLATGPFFDD73 pKa = 3.94ATSPTVAGGKK83 pKa = 9.28YY84 pKa = 10.19LYY86 pKa = 10.83LEE88 pKa = 4.39TSGGALNDD96 pKa = 3.55QNTLSSPPVSIPMSATGVKK115 pKa = 10.23LDD117 pKa = 3.91FAYY120 pKa = 10.78HH121 pKa = 5.5MFGASMGEE129 pKa = 4.08LYY131 pKa = 10.97VVIDD135 pKa = 3.97TNNVSDD141 pKa = 4.16TVVTLIGQQQTSGADD156 pKa = 3.42PFITSSTILTGYY168 pKa = 9.59AGKK171 pKa = 8.28TLTVKK176 pKa = 10.68FIGKK180 pKa = 9.85RR181 pKa = 11.84GTSYY185 pKa = 10.5TGDD188 pKa = 3.09ISIDD192 pKa = 3.67EE193 pKa = 4.5VKK195 pKa = 10.64LFQPVPNEE203 pKa = 3.74MGITEE208 pKa = 4.22ITTPTSSCGLTASEE222 pKa = 4.48VVTIEE227 pKa = 3.82VQNFGTSNITSFDD240 pKa = 3.15ASFSINGGTPVTEE253 pKa = 4.34TVTTTITPNSIYY265 pKa = 10.33SYY267 pKa = 10.23TFTATANLSAFGPYY281 pKa = 8.35EE282 pKa = 3.93VKK284 pKa = 10.71SYY286 pKa = 9.3VTLLNDD292 pKa = 3.59PTSSNDD298 pKa = 3.56TLVKK302 pKa = 10.67NITNIPILNGFPYY315 pKa = 10.98LEE317 pKa = 4.35TFEE320 pKa = 5.02SGPGGWISGGTNSSWALGTPAGTVINSAAGGTNSWVTNLTGTYY363 pKa = 9.72NATEE367 pKa = 3.81ASYY370 pKa = 11.27VVSPCFDD377 pKa = 3.53FTSLVAPVFSADD389 pKa = 3.31VWWNSEE395 pKa = 3.92FSWDD399 pKa = 3.51GAILQTSIDD408 pKa = 5.21GGTTWQKK415 pKa = 10.93VGANGDD421 pKa = 3.78PNNWYY426 pKa = 8.91TDD428 pKa = 3.48NTISGLTPLAPSQEE442 pKa = 4.01GWTGRR447 pKa = 11.84NSTSNGSGGWVKK459 pKa = 10.63VKK461 pKa = 10.94APLTGLGSVSGVILRR476 pKa = 11.84IAFGSDD482 pKa = 2.8GSVMDD487 pKa = 4.46EE488 pKa = 4.63GFAFDD493 pKa = 5.37NIQIYY498 pKa = 7.61DD499 pKa = 3.89TPAQDD504 pKa = 5.29AEE506 pKa = 4.5LLSITSPINGCGLTSTEE523 pKa = 3.88QAEE526 pKa = 4.41VQIVNAGSASISNFPVSITVNGGTPLTEE554 pKa = 4.07TVTATILPLDD564 pKa = 3.57TLFYY568 pKa = 10.48TFTGTVNLSTTGNYY582 pKa = 9.82SIKK585 pKa = 10.36AYY587 pKa = 9.67TGLTGDD593 pKa = 4.51GLSSNDD599 pKa = 3.2TATKK603 pKa = 10.25VVTHH607 pKa = 6.6IPIISTFPYY616 pKa = 8.68STSFEE621 pKa = 4.14NSNEE625 pKa = 3.69GWLSSGINNSWSLGVPAGSIIDD647 pKa = 3.76TASNGTKK654 pKa = 10.2AWVTNLNGVYY664 pKa = 10.59NNNEE668 pKa = 3.57QSFVVSPCLDD678 pKa = 3.58FTNLTSPLISLDD690 pKa = 3.15IQYY693 pKa = 10.99EE694 pKa = 4.15LDD696 pKa = 3.48NSNDD700 pKa = 3.32GVVLQSSIDD709 pKa = 4.49GGATWQKK716 pKa = 10.43VGAFGSPNNWYY727 pKa = 8.87TDD729 pKa = 3.37NTIVGLSSIEE739 pKa = 3.95TSQEE743 pKa = 3.12GWTGSGTSGSNGWKK757 pKa = 9.51SARR760 pKa = 11.84AALSGLGGEE769 pKa = 4.38PSVILRR775 pKa = 11.84IALGTNGFTTFEE787 pKa = 4.07GFAFDD792 pKa = 4.18NVTIQEE798 pKa = 4.43APSTDD803 pKa = 3.16AGVVDD808 pKa = 5.68IIDD811 pKa = 4.08PASGCGLSSAEE822 pKa = 4.12VVTVRR827 pKa = 11.84IVNDD831 pKa = 3.62GATNISNFPVSVVLNNGTPLTEE853 pKa = 4.2TVTATMLPNDD863 pKa = 3.71TLTYY867 pKa = 10.32TFTGTLNMSALGTYY881 pKa = 10.1SVVSYY886 pKa = 10.53TSLPLDD892 pKa = 3.59GFNVNDD898 pKa = 4.0TASKK902 pKa = 8.75TVVNIPVISGFPYY915 pKa = 9.56TEE917 pKa = 4.08SFEE920 pKa = 4.28TSAGGWTVDD929 pKa = 3.24GGTSSFALGAPIGSLINSASNGTQAWVTNLSGNYY963 pKa = 9.49LADD966 pKa = 3.23EE967 pKa = 4.58AGYY970 pKa = 10.77VKK972 pKa = 10.55SPCLDD977 pKa = 3.8FSTLIAPIISMDD989 pKa = 3.44VWWNSEE995 pKa = 3.95FSWDD999 pKa = 3.6GAVLQSSIDD1008 pKa = 4.72GGTTWQKK1015 pKa = 10.65VGSFGDD1021 pKa = 3.58PNNWYY1026 pKa = 10.18NDD1028 pKa = 3.16NSINGLNTGSPILEE1042 pKa = 4.11PSGEE1046 pKa = 3.97GWTGRR1051 pKa = 11.84NSSSNGSNGWVNATHH1066 pKa = 6.51QLTGLGGQPGVIFRR1080 pKa = 11.84VAFGSDD1086 pKa = 3.14GSVMDD1091 pKa = 4.46EE1092 pKa = 4.54GFAFDD1097 pKa = 3.9NVQIYY1102 pKa = 7.96EE1103 pKa = 4.31SPAVDD1108 pKa = 3.61IKK1110 pKa = 11.43AMEE1113 pKa = 4.05VSRR1116 pKa = 11.84PLVGCGLSATEE1127 pKa = 4.48TIAARR1132 pKa = 11.84YY1133 pKa = 7.53EE1134 pKa = 4.02NLGTDD1139 pKa = 3.42TLTNFPVAYY1148 pKa = 9.39SVNGVAITPEE1158 pKa = 4.3TFTDD1162 pKa = 3.59TLFPGDD1168 pKa = 3.45KK1169 pKa = 9.91KK1170 pKa = 11.09SYY1172 pKa = 10.65EE1173 pKa = 4.14FTTTANLSSTIQYY1186 pKa = 9.18TIEE1189 pKa = 3.93VWSAAAGDD1197 pKa = 4.9AITSNDD1203 pKa = 3.54TASVTFTNIAASTNPYY1219 pKa = 9.13PQTFDD1224 pKa = 3.48VLTDD1228 pKa = 3.71GATDD1232 pKa = 3.85FSPINWTPINAGAFDD1247 pKa = 3.72WRR1249 pKa = 11.84AEE1251 pKa = 4.15TT1252 pKa = 4.54
Molecular weight: 131.17 kDa
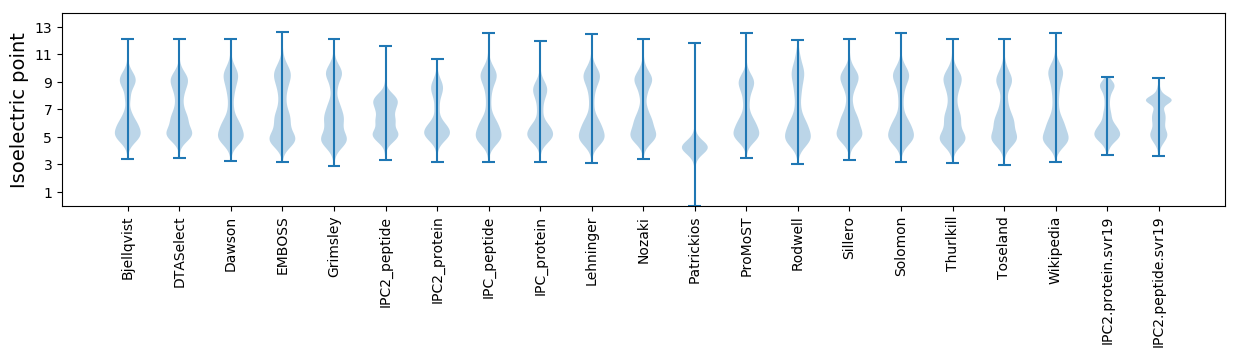
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6N9NJ49|A0A6N9NJ49_9FLAO DUF4834 family protein OS=Acidiluteibacter ferrifornacis OX=2692424 GN=GQN54_07115 PE=4 SV=1
MM1 pKa = 7.19IQQEE5 pKa = 4.43SRR7 pKa = 11.84LSVADD12 pKa = 3.32NSGAKK17 pKa = 9.22EE18 pKa = 3.93VLCIRR23 pKa = 11.84VLGGTGKK30 pKa = 10.44RR31 pKa = 11.84YY32 pKa = 10.3ARR34 pKa = 11.84IGDD37 pKa = 3.98KK38 pKa = 10.52IVVSVKK44 pKa = 10.53SAIPSGNIKK53 pKa = 10.12KK54 pKa = 10.34GAVSKK59 pKa = 10.89AVVVRR64 pKa = 11.84TRR66 pKa = 11.84KK67 pKa = 9.57EE68 pKa = 3.23IRR70 pKa = 11.84RR71 pKa = 11.84PDD73 pKa = 3.16GSYY76 pKa = 10.62IRR78 pKa = 11.84FDD80 pKa = 3.58NNAVVLLNAAGEE92 pKa = 4.21MRR94 pKa = 11.84GTRR97 pKa = 11.84IFGPVARR104 pKa = 11.84EE105 pKa = 3.54LRR107 pKa = 11.84DD108 pKa = 3.52RR109 pKa = 11.84QFMKK113 pKa = 10.24IVSLAPEE120 pKa = 4.12VLL122 pKa = 3.56
MM1 pKa = 7.19IQQEE5 pKa = 4.43SRR7 pKa = 11.84LSVADD12 pKa = 3.32NSGAKK17 pKa = 9.22EE18 pKa = 3.93VLCIRR23 pKa = 11.84VLGGTGKK30 pKa = 10.44RR31 pKa = 11.84YY32 pKa = 10.3ARR34 pKa = 11.84IGDD37 pKa = 3.98KK38 pKa = 10.52IVVSVKK44 pKa = 10.53SAIPSGNIKK53 pKa = 10.12KK54 pKa = 10.34GAVSKK59 pKa = 10.89AVVVRR64 pKa = 11.84TRR66 pKa = 11.84KK67 pKa = 9.57EE68 pKa = 3.23IRR70 pKa = 11.84RR71 pKa = 11.84PDD73 pKa = 3.16GSYY76 pKa = 10.62IRR78 pKa = 11.84FDD80 pKa = 3.58NNAVVLLNAAGEE92 pKa = 4.21MRR94 pKa = 11.84GTRR97 pKa = 11.84IFGPVARR104 pKa = 11.84EE105 pKa = 3.54LRR107 pKa = 11.84DD108 pKa = 3.52RR109 pKa = 11.84QFMKK113 pKa = 10.24IVSLAPEE120 pKa = 4.12VLL122 pKa = 3.56
Molecular weight: 13.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1071256 |
27 |
3996 |
355.9 |
40.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.337 ± 0.056 | 0.793 ± 0.019 |
5.372 ± 0.041 | 6.467 ± 0.056 |
5.236 ± 0.041 | 6.461 ± 0.052 |
1.686 ± 0.021 | 8.146 ± 0.045 |
7.221 ± 0.067 | 9.23 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.157 ± 0.025 | 6.216 ± 0.053 |
3.405 ± 0.028 | 3.526 ± 0.029 |
3.423 ± 0.032 | 7.172 ± 0.049 |
5.699 ± 0.077 | 6.205 ± 0.038 |
1.051 ± 0.019 | 4.196 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
