
Breoghania sp. L-A4
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Breoghaniaceae; Breoghania; unclassified Breoghania
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
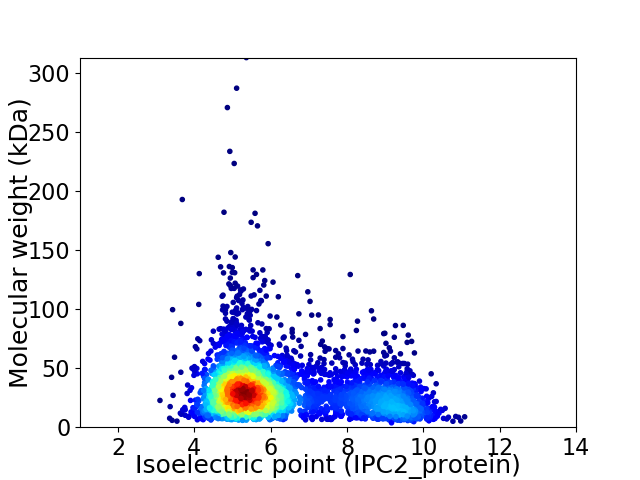
Virtual 2D-PAGE plot for 4084 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346R4X8|A0A346R4X8_9RHIZ GntR family transcriptional regulator OS=Breoghania sp. L-A4 OX=2304600 GN=D1F64_17895 PE=4 SV=1
MM1 pKa = 7.6SGTGNTTKK9 pKa = 10.59SVASTGNQFIDD20 pKa = 4.17GVLTYY25 pKa = 10.33DD26 pKa = 3.23AWADD30 pKa = 3.31STLYY34 pKa = 10.94YY35 pKa = 10.78SFSTSSAAYY44 pKa = 9.93SYY46 pKa = 10.68TGTGNEE52 pKa = 4.3DD53 pKa = 3.05LPANFNAITAAQQTAAHH70 pKa = 6.59FALSADD76 pKa = 4.49DD77 pKa = 5.14GPSASAGFSLEE88 pKa = 3.68GFTAINVASLGSTNSSTAEE107 pKa = 3.45IRR109 pKa = 11.84FGEE112 pKa = 4.43TSSSSLGTAQVADD125 pKa = 4.27FPGNYY130 pKa = 7.48ITGPSGDD137 pKa = 4.4NGDD140 pKa = 3.46VWFGSYY146 pKa = 9.84SGYY149 pKa = 10.42IYY151 pKa = 8.17RR152 pKa = 11.84TPVAGNYY159 pKa = 8.57AWHH162 pKa = 6.05THH164 pKa = 4.24LHH166 pKa = 6.28EE167 pKa = 5.26IGHH170 pKa = 6.46ALGLKK175 pKa = 9.87HH176 pKa = 6.06GQEE179 pKa = 4.22NSGAGPTPAAYY190 pKa = 10.21DD191 pKa = 3.51SMEE194 pKa = 4.2YY195 pKa = 10.89SVMTYY200 pKa = 9.8RR201 pKa = 11.84SYY203 pKa = 12.09VNDD206 pKa = 4.26PLIGGYY212 pKa = 9.39SNEE215 pKa = 4.35TYY217 pKa = 10.61GYY219 pKa = 8.22AQSYY223 pKa = 8.49MMADD227 pKa = 3.49IAALQHH233 pKa = 6.14MYY235 pKa = 10.79GADD238 pKa = 3.36FTTNSGDD245 pKa = 3.86TVYY248 pKa = 10.64KK249 pKa = 10.01WNPNSGDD256 pKa = 3.6TLVNGAVAIDD266 pKa = 3.47AGGNRR271 pKa = 11.84IFATVWDD278 pKa = 4.19GGGTDD283 pKa = 3.72TYY285 pKa = 11.43DD286 pKa = 3.09LSSYY290 pKa = 7.54TTALMLDD297 pKa = 4.3LRR299 pKa = 11.84PGEE302 pKa = 4.25HH303 pKa = 6.36SVFSAVQLAYY313 pKa = 10.78LGDD316 pKa = 3.88GNYY319 pKa = 10.31SRR321 pKa = 11.84GNIFNALQYY330 pKa = 10.93QSDD333 pKa = 3.79TRR335 pKa = 11.84SLIEE339 pKa = 3.81NAIGGSGDD347 pKa = 3.52DD348 pKa = 4.24TITGNAANNTLQGGNGADD366 pKa = 3.45MLYY369 pKa = 10.97GGLGNDD375 pKa = 3.72TLEE378 pKa = 4.69GGAGADD384 pKa = 4.01LLDD387 pKa = 4.56GQGGWDD393 pKa = 3.33MASYY397 pKa = 10.13AGSSAAVTVNLGDD410 pKa = 3.94GLTEE414 pKa = 3.98TGGDD418 pKa = 3.56AEE420 pKa = 4.61GDD422 pKa = 3.24QLLRR426 pKa = 11.84IEE428 pKa = 4.5HH429 pKa = 6.56LEE431 pKa = 3.9GSVFHH436 pKa = 7.18DD437 pKa = 3.57TLIGNAGINYY447 pKa = 9.84LYY449 pKa = 10.62GGNGNDD455 pKa = 3.72TLEE458 pKa = 4.62GGAGADD464 pKa = 4.01LLDD467 pKa = 4.7GQGGWDD473 pKa = 3.23RR474 pKa = 11.84ASYY477 pKa = 10.1AGSSAAVTVNLGDD490 pKa = 3.94GLTEE494 pKa = 3.98TGGDD498 pKa = 3.59AEE500 pKa = 4.83GDD502 pKa = 3.38LLLRR506 pKa = 11.84IEE508 pKa = 4.57HH509 pKa = 6.75LEE511 pKa = 4.15GSAFDD516 pKa = 3.78DD517 pKa = 3.94TLIGSAGINYY527 pKa = 10.04LYY529 pKa = 10.98GGDD532 pKa = 4.25GNDD535 pKa = 3.66TLEE538 pKa = 4.65GGAGADD544 pKa = 3.65RR545 pKa = 11.84LDD547 pKa = 4.27GQGGWDD553 pKa = 3.18RR554 pKa = 11.84ASYY557 pKa = 9.01TGSSAAVTVNLGDD570 pKa = 3.85GLVEE574 pKa = 4.18SGGDD578 pKa = 3.49AEE580 pKa = 5.25GDD582 pKa = 3.18QLLRR586 pKa = 11.84IEE588 pKa = 4.66HH589 pKa = 6.58LEE591 pKa = 4.04GSAFHH596 pKa = 6.97DD597 pKa = 3.86TLIGSAGINYY607 pKa = 10.04LYY609 pKa = 10.98GGDD612 pKa = 4.25GNDD615 pKa = 3.66TLEE618 pKa = 4.65GGAGADD624 pKa = 3.65RR625 pKa = 11.84LDD627 pKa = 4.21GQGGWDD633 pKa = 3.33MASYY637 pKa = 10.13AGSSAAVSVNLGDD650 pKa = 4.12GLVEE654 pKa = 4.18SGGDD658 pKa = 3.5AEE660 pKa = 4.7GDD662 pKa = 3.06RR663 pKa = 11.84LLRR666 pKa = 11.84IEE668 pKa = 4.6HH669 pKa = 6.72LEE671 pKa = 4.15GSAFSDD677 pKa = 3.59TLIGSAGINYY687 pKa = 9.51LFGGDD692 pKa = 4.18GNDD695 pKa = 3.65TLEE698 pKa = 4.65GGAGADD704 pKa = 3.65RR705 pKa = 11.84LDD707 pKa = 4.52GQGGWDD713 pKa = 3.21KK714 pKa = 11.25VSYY717 pKa = 9.8AGSSAGVSVNLGDD730 pKa = 4.24GLVEE734 pKa = 4.18SGGDD738 pKa = 3.49AEE740 pKa = 5.25GDD742 pKa = 3.18QLLRR746 pKa = 11.84IEE748 pKa = 4.42HH749 pKa = 6.8LEE751 pKa = 4.17GSSYY755 pKa = 11.72ADD757 pKa = 3.27ILTGSAGINWMGGGAGADD775 pKa = 3.58EE776 pKa = 4.3LAGGGGNDD784 pKa = 3.62RR785 pKa = 11.84LSGGTGADD793 pKa = 3.03DD794 pKa = 5.38FIFNTGDD801 pKa = 3.37GSDD804 pKa = 3.89TITDD808 pKa = 4.39FEE810 pKa = 5.85LGLDD814 pKa = 4.5DD815 pKa = 5.64IRR817 pKa = 11.84ILSGASSFGDD827 pKa = 3.41LTITDD832 pKa = 3.8SGANALVQFADD843 pKa = 3.99VSITLLNVDD852 pKa = 4.21HH853 pKa = 6.95TLLMSDD859 pKa = 4.73DD860 pKa = 4.06FLFAA864 pKa = 5.48
MM1 pKa = 7.6SGTGNTTKK9 pKa = 10.59SVASTGNQFIDD20 pKa = 4.17GVLTYY25 pKa = 10.33DD26 pKa = 3.23AWADD30 pKa = 3.31STLYY34 pKa = 10.94YY35 pKa = 10.78SFSTSSAAYY44 pKa = 9.93SYY46 pKa = 10.68TGTGNEE52 pKa = 4.3DD53 pKa = 3.05LPANFNAITAAQQTAAHH70 pKa = 6.59FALSADD76 pKa = 4.49DD77 pKa = 5.14GPSASAGFSLEE88 pKa = 3.68GFTAINVASLGSTNSSTAEE107 pKa = 3.45IRR109 pKa = 11.84FGEE112 pKa = 4.43TSSSSLGTAQVADD125 pKa = 4.27FPGNYY130 pKa = 7.48ITGPSGDD137 pKa = 4.4NGDD140 pKa = 3.46VWFGSYY146 pKa = 9.84SGYY149 pKa = 10.42IYY151 pKa = 8.17RR152 pKa = 11.84TPVAGNYY159 pKa = 8.57AWHH162 pKa = 6.05THH164 pKa = 4.24LHH166 pKa = 6.28EE167 pKa = 5.26IGHH170 pKa = 6.46ALGLKK175 pKa = 9.87HH176 pKa = 6.06GQEE179 pKa = 4.22NSGAGPTPAAYY190 pKa = 10.21DD191 pKa = 3.51SMEE194 pKa = 4.2YY195 pKa = 10.89SVMTYY200 pKa = 9.8RR201 pKa = 11.84SYY203 pKa = 12.09VNDD206 pKa = 4.26PLIGGYY212 pKa = 9.39SNEE215 pKa = 4.35TYY217 pKa = 10.61GYY219 pKa = 8.22AQSYY223 pKa = 8.49MMADD227 pKa = 3.49IAALQHH233 pKa = 6.14MYY235 pKa = 10.79GADD238 pKa = 3.36FTTNSGDD245 pKa = 3.86TVYY248 pKa = 10.64KK249 pKa = 10.01WNPNSGDD256 pKa = 3.6TLVNGAVAIDD266 pKa = 3.47AGGNRR271 pKa = 11.84IFATVWDD278 pKa = 4.19GGGTDD283 pKa = 3.72TYY285 pKa = 11.43DD286 pKa = 3.09LSSYY290 pKa = 7.54TTALMLDD297 pKa = 4.3LRR299 pKa = 11.84PGEE302 pKa = 4.25HH303 pKa = 6.36SVFSAVQLAYY313 pKa = 10.78LGDD316 pKa = 3.88GNYY319 pKa = 10.31SRR321 pKa = 11.84GNIFNALQYY330 pKa = 10.93QSDD333 pKa = 3.79TRR335 pKa = 11.84SLIEE339 pKa = 3.81NAIGGSGDD347 pKa = 3.52DD348 pKa = 4.24TITGNAANNTLQGGNGADD366 pKa = 3.45MLYY369 pKa = 10.97GGLGNDD375 pKa = 3.72TLEE378 pKa = 4.69GGAGADD384 pKa = 4.01LLDD387 pKa = 4.56GQGGWDD393 pKa = 3.33MASYY397 pKa = 10.13AGSSAAVTVNLGDD410 pKa = 3.94GLTEE414 pKa = 3.98TGGDD418 pKa = 3.56AEE420 pKa = 4.61GDD422 pKa = 3.24QLLRR426 pKa = 11.84IEE428 pKa = 4.5HH429 pKa = 6.56LEE431 pKa = 3.9GSVFHH436 pKa = 7.18DD437 pKa = 3.57TLIGNAGINYY447 pKa = 9.84LYY449 pKa = 10.62GGNGNDD455 pKa = 3.72TLEE458 pKa = 4.62GGAGADD464 pKa = 4.01LLDD467 pKa = 4.7GQGGWDD473 pKa = 3.23RR474 pKa = 11.84ASYY477 pKa = 10.1AGSSAAVTVNLGDD490 pKa = 3.94GLTEE494 pKa = 3.98TGGDD498 pKa = 3.59AEE500 pKa = 4.83GDD502 pKa = 3.38LLLRR506 pKa = 11.84IEE508 pKa = 4.57HH509 pKa = 6.75LEE511 pKa = 4.15GSAFDD516 pKa = 3.78DD517 pKa = 3.94TLIGSAGINYY527 pKa = 10.04LYY529 pKa = 10.98GGDD532 pKa = 4.25GNDD535 pKa = 3.66TLEE538 pKa = 4.65GGAGADD544 pKa = 3.65RR545 pKa = 11.84LDD547 pKa = 4.27GQGGWDD553 pKa = 3.18RR554 pKa = 11.84ASYY557 pKa = 9.01TGSSAAVTVNLGDD570 pKa = 3.85GLVEE574 pKa = 4.18SGGDD578 pKa = 3.49AEE580 pKa = 5.25GDD582 pKa = 3.18QLLRR586 pKa = 11.84IEE588 pKa = 4.66HH589 pKa = 6.58LEE591 pKa = 4.04GSAFHH596 pKa = 6.97DD597 pKa = 3.86TLIGSAGINYY607 pKa = 10.04LYY609 pKa = 10.98GGDD612 pKa = 4.25GNDD615 pKa = 3.66TLEE618 pKa = 4.65GGAGADD624 pKa = 3.65RR625 pKa = 11.84LDD627 pKa = 4.21GQGGWDD633 pKa = 3.33MASYY637 pKa = 10.13AGSSAAVSVNLGDD650 pKa = 4.12GLVEE654 pKa = 4.18SGGDD658 pKa = 3.5AEE660 pKa = 4.7GDD662 pKa = 3.06RR663 pKa = 11.84LLRR666 pKa = 11.84IEE668 pKa = 4.6HH669 pKa = 6.72LEE671 pKa = 4.15GSAFSDD677 pKa = 3.59TLIGSAGINYY687 pKa = 9.51LFGGDD692 pKa = 4.18GNDD695 pKa = 3.65TLEE698 pKa = 4.65GGAGADD704 pKa = 3.65RR705 pKa = 11.84LDD707 pKa = 4.52GQGGWDD713 pKa = 3.21KK714 pKa = 11.25VSYY717 pKa = 9.8AGSSAGVSVNLGDD730 pKa = 4.24GLVEE734 pKa = 4.18SGGDD738 pKa = 3.49AEE740 pKa = 5.25GDD742 pKa = 3.18QLLRR746 pKa = 11.84IEE748 pKa = 4.42HH749 pKa = 6.8LEE751 pKa = 4.17GSSYY755 pKa = 11.72ADD757 pKa = 3.27ILTGSAGINWMGGGAGADD775 pKa = 3.58EE776 pKa = 4.3LAGGGGNDD784 pKa = 3.62RR785 pKa = 11.84LSGGTGADD793 pKa = 3.03DD794 pKa = 5.38FIFNTGDD801 pKa = 3.37GSDD804 pKa = 3.89TITDD808 pKa = 4.39FEE810 pKa = 5.85LGLDD814 pKa = 4.5DD815 pKa = 5.64IRR817 pKa = 11.84ILSGASSFGDD827 pKa = 3.41LTITDD832 pKa = 3.8SGANALVQFADD843 pKa = 3.99VSITLLNVDD852 pKa = 4.21HH853 pKa = 6.95TLLMSDD859 pKa = 4.73DD860 pKa = 4.06FLFAA864 pKa = 5.48
Molecular weight: 87.99 kDa
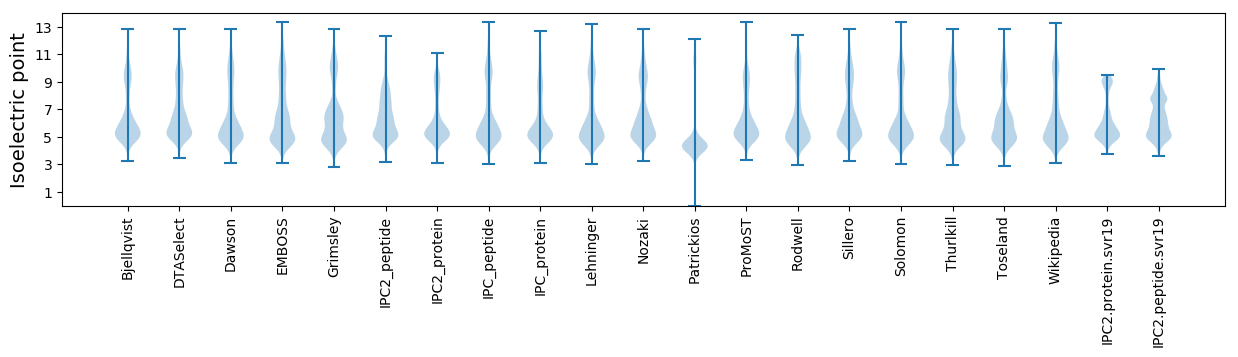
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346QXF9|A0A346QXF9_9RHIZ Heme exporter protein D OS=Breoghania sp. L-A4 OX=2304600 GN=ccmD PE=3 SV=1
MM1 pKa = 6.76WTRR4 pKa = 11.84IFISVLVFMQINAVLFGAGIVAVLVVPALKK34 pKa = 10.19AQAMMLLPAVVVASLVIALPVSWWLAPRR62 pKa = 11.84LRR64 pKa = 11.84LPRR67 pKa = 11.84WRR69 pKa = 11.84RR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84HH73 pKa = 5.23AANAGQQ79 pKa = 3.38
MM1 pKa = 6.76WTRR4 pKa = 11.84IFISVLVFMQINAVLFGAGIVAVLVVPALKK34 pKa = 10.19AQAMMLLPAVVVASLVIALPVSWWLAPRR62 pKa = 11.84LRR64 pKa = 11.84LPRR67 pKa = 11.84WRR69 pKa = 11.84RR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84HH73 pKa = 5.23AANAGQQ79 pKa = 3.38
Molecular weight: 8.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1258513 |
33 |
2978 |
308.2 |
33.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.67 ± 0.052 | 0.922 ± 0.013 |
6.034 ± 0.031 | 5.731 ± 0.033 |
3.726 ± 0.024 | 8.624 ± 0.038 |
2.049 ± 0.017 | 5.303 ± 0.029 |
3.123 ± 0.028 | 9.897 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.574 ± 0.02 | 2.544 ± 0.019 |
5.057 ± 0.028 | 2.904 ± 0.024 |
7.075 ± 0.041 | 5.442 ± 0.028 |
5.47 ± 0.031 | 7.39 ± 0.035 |
1.284 ± 0.013 | 2.185 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
