
Changjiang sobemo-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.74
Get precalculated fractions of proteins
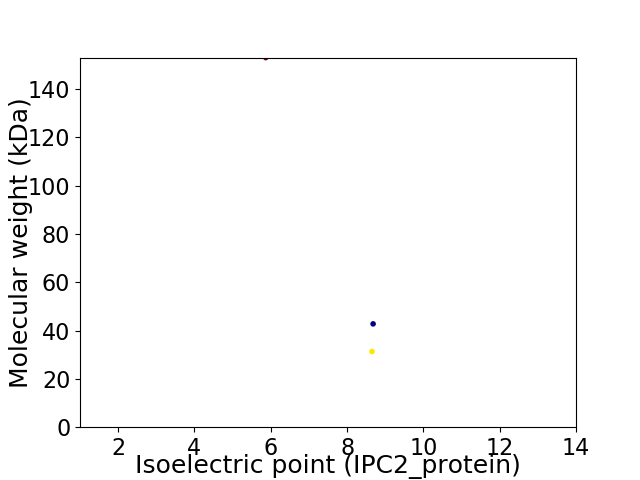
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
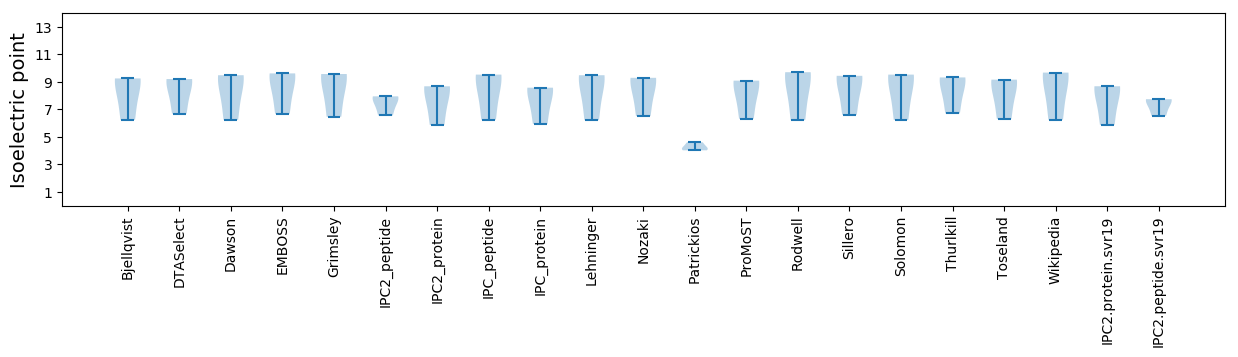
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEG8|A0A1L3KEG8_9VIRU Capsid protein OS=Changjiang sobemo-like virus 2 OX=1922801 PE=3 SV=1
MM1 pKa = 7.6IVPKK5 pKa = 10.85NKK7 pKa = 9.32FCQSDD12 pKa = 3.71DD13 pKa = 3.41YY14 pKa = 11.57DD15 pKa = 3.58YY16 pKa = 11.52CSRR19 pKa = 11.84DD20 pKa = 3.46KK21 pKa = 11.17CWVCLIPLRR30 pKa = 11.84CSDD33 pKa = 5.26LIEE36 pKa = 5.01KK37 pKa = 9.66YY38 pKa = 10.14ISYY41 pKa = 10.61DD42 pKa = 3.08PYY44 pKa = 10.38EE45 pKa = 4.56AFGSEE50 pKa = 4.47YY51 pKa = 10.62EE52 pKa = 4.63SIVLPCEE59 pKa = 3.58RR60 pKa = 11.84LAALCYY66 pKa = 10.45YY67 pKa = 9.99EE68 pKa = 5.11SLWTEE73 pKa = 3.9VRR75 pKa = 11.84RR76 pKa = 11.84SSTEE80 pKa = 3.24WSIARR85 pKa = 11.84WYY87 pKa = 8.62STAVDD92 pKa = 3.21HH93 pKa = 7.26RR94 pKa = 11.84GRR96 pKa = 11.84PRR98 pKa = 11.84AFRR101 pKa = 11.84IVGTNLHH108 pKa = 6.77DD109 pKa = 4.27LAGVAAMPYY118 pKa = 8.65PPTMSEE124 pKa = 3.58MEE126 pKa = 4.62YY127 pKa = 10.2ILKK130 pKa = 9.81EE131 pKa = 4.21VPHH134 pKa = 7.29DD135 pKa = 4.98DD136 pKa = 3.7IPGEE140 pKa = 4.08VFKK143 pKa = 11.39YY144 pKa = 10.49LGGRR148 pKa = 11.84TKK150 pKa = 11.02LSFTLKK156 pKa = 10.05LFLMFGVLSGTIFSVEE172 pKa = 4.05ASDD175 pKa = 5.17QYY177 pKa = 11.61TSDD180 pKa = 3.43LVNTGEE186 pKa = 4.23LARR189 pKa = 11.84EE190 pKa = 3.78ITLWPVNEE198 pKa = 4.15SFRR201 pKa = 11.84MLEE204 pKa = 3.87HH205 pKa = 5.76TTISTTQCHH214 pKa = 5.8YY215 pKa = 11.48EE216 pKa = 3.85NGTCVEE222 pKa = 4.23YY223 pKa = 10.41PNLVNQVFDD232 pKa = 3.62WLRR235 pKa = 11.84VRR237 pKa = 11.84VRR239 pKa = 11.84NLCYY243 pKa = 10.63LLDD246 pKa = 4.23EE247 pKa = 4.54STLLARR253 pKa = 11.84HH254 pKa = 5.95SSSVAKK260 pKa = 10.26EE261 pKa = 3.37RR262 pKa = 11.84TRR264 pKa = 11.84FYY266 pKa = 11.47KK267 pKa = 10.71NIIVKK272 pKa = 10.22DD273 pKa = 3.79DD274 pKa = 3.66YY275 pKa = 11.47FRR277 pKa = 11.84AEE279 pKa = 4.09YY280 pKa = 10.89DD281 pKa = 4.21DD282 pKa = 5.07ISTAEE287 pKa = 3.89NEE289 pKa = 4.75LYY291 pKa = 11.03NFMMLFLSFLYY302 pKa = 10.42GINSGFWSLCILFINFLYY320 pKa = 10.41FMRR323 pKa = 11.84FAHH326 pKa = 6.02NFVVSTPLGFWIYY339 pKa = 10.54TKK341 pKa = 10.19VVWVLGIWLGYY352 pKa = 8.78YY353 pKa = 8.12VTYY356 pKa = 10.15HH357 pKa = 6.59ALVVLLAASISYY369 pKa = 9.23LHH371 pKa = 6.72SKK373 pKa = 9.63MIEE376 pKa = 3.94LAVVVVMAPYY386 pKa = 9.94HH387 pKa = 4.78IWSRR391 pKa = 11.84FRR393 pKa = 11.84RR394 pKa = 11.84TGVKK398 pKa = 10.04GFVRR402 pKa = 11.84EE403 pKa = 4.13MILPGSKK410 pKa = 8.86IEE412 pKa = 4.16KK413 pKa = 10.2FIGAKK418 pKa = 9.31QPAVAALVVEE428 pKa = 4.94VDD430 pKa = 4.22GKK432 pKa = 7.88WHH434 pKa = 5.8YY435 pKa = 11.43SAMCFRR441 pKa = 11.84YY442 pKa = 9.86EE443 pKa = 3.85NHH445 pKa = 6.48LVSAFHH451 pKa = 7.01PFAKK455 pKa = 10.12HH456 pKa = 4.43MSVGRR461 pKa = 11.84RR462 pKa = 11.84MGVAPNLLCKK472 pKa = 10.33KK473 pKa = 9.83RR474 pKa = 11.84WTHH477 pKa = 6.81DD478 pKa = 3.32FGAIQVIDD486 pKa = 3.97PSNYY490 pKa = 7.98IVDD493 pKa = 4.53FEE495 pKa = 4.34RR496 pKa = 11.84HH497 pKa = 5.58DD498 pKa = 3.7VFMLHH503 pKa = 6.27MSGEE507 pKa = 3.78FWDD510 pKa = 4.01RR511 pKa = 11.84VRR513 pKa = 11.84LPEE516 pKa = 4.18PKK518 pKa = 10.07KK519 pKa = 10.74SVSNSFNDD527 pKa = 3.32SATVFSYY534 pKa = 10.17MAGTLHH540 pKa = 6.6TSNGVLTSHH549 pKa = 6.78TNVDD553 pKa = 3.66YY554 pKa = 10.68PPNNFMAHH562 pKa = 5.43TCSTLPGFSGTPVFAANGAVRR583 pKa = 11.84GVHH586 pKa = 6.11IGSDD590 pKa = 3.33NYY592 pKa = 10.31EE593 pKa = 3.83EE594 pKa = 4.36RR595 pKa = 11.84NIMFRR600 pKa = 11.84IDD602 pKa = 3.45HH603 pKa = 6.32IKK605 pKa = 10.93YY606 pKa = 9.55YY607 pKa = 10.76LNKK610 pKa = 10.43NKK612 pKa = 8.07VTRR615 pKa = 11.84EE616 pKa = 3.98DD617 pKa = 3.35SSDD620 pKa = 3.56FQKK623 pKa = 11.08KK624 pKa = 7.11VLRR627 pKa = 11.84SMKK630 pKa = 9.64RR631 pKa = 11.84RR632 pKa = 11.84KK633 pKa = 9.86GKK635 pKa = 9.17EE636 pKa = 3.24AVYY639 pKa = 10.9YY640 pKa = 10.73FDD642 pKa = 6.44DD643 pKa = 4.25FDD645 pKa = 4.56EE646 pKa = 5.43DD647 pKa = 3.56NSVAYY652 pKa = 9.86DD653 pKa = 3.22IRR655 pKa = 11.84TGQIEE660 pKa = 4.27RR661 pKa = 11.84VPRR664 pKa = 11.84TEE666 pKa = 4.36EE667 pKa = 3.54EE668 pKa = 4.67DD669 pKa = 3.6EE670 pKa = 4.32EE671 pKa = 4.37FWRR674 pKa = 11.84VEE676 pKa = 3.71MQKK679 pKa = 10.5PVGLWGDD686 pKa = 3.84YY687 pKa = 10.65EE688 pKa = 4.26DD689 pKa = 5.47APISKK694 pKa = 10.09EE695 pKa = 3.94EE696 pKa = 4.51EE697 pKa = 3.73IDD699 pKa = 3.65EE700 pKa = 4.39VEE702 pKa = 3.9PSQFFEE708 pKa = 4.17VLDD711 pKa = 3.73MRR713 pKa = 11.84KK714 pKa = 9.37KK715 pKa = 10.31LHH717 pKa = 6.71SPTPDD722 pKa = 2.93TKK724 pKa = 10.65HH725 pKa = 6.98AEE727 pKa = 4.08TTEE730 pKa = 3.76ILEE733 pKa = 4.17PFMDD737 pKa = 3.63KK738 pKa = 10.56FKK740 pKa = 10.99EE741 pKa = 3.92LGYY744 pKa = 10.13VEE746 pKa = 6.25GMFEE750 pKa = 4.06YY751 pKa = 9.84PAAGHH756 pKa = 6.93KK757 pKa = 9.42MDD759 pKa = 4.61TKK761 pKa = 10.85SCVNAIIQHH770 pKa = 4.94GTNVSEE776 pKa = 4.85CPVPITPDD784 pKa = 3.24EE785 pKa = 4.14EE786 pKa = 4.54DD787 pKa = 3.41RR788 pKa = 11.84VVDD791 pKa = 3.81AVSAMVQEE799 pKa = 4.87FSFVVDD805 pKa = 3.87PNYY808 pKa = 10.83KK809 pKa = 9.1SLEE812 pKa = 4.02YY813 pKa = 10.55FEE815 pKa = 6.44SIINTNQIEE824 pKa = 4.27DD825 pKa = 3.72RR826 pKa = 11.84KK827 pKa = 10.24SPGMPYY833 pKa = 10.21LEE835 pKa = 4.51EE836 pKa = 5.27GMTTNAQVLDD846 pKa = 3.9KK847 pKa = 11.51LGVTWLYY854 pKa = 11.74NKK856 pKa = 9.83MIEE859 pKa = 4.29CWNMSILYY867 pKa = 10.42RR868 pKa = 11.84MFLKK872 pKa = 10.98YY873 pKa = 10.63EE874 pKa = 3.8PTKK877 pKa = 10.55KK878 pKa = 10.43KK879 pKa = 10.83KK880 pKa = 9.31IDD882 pKa = 2.84NWMARR887 pKa = 11.84VILSFPLHH895 pKa = 6.0KK896 pKa = 9.46TLQNIVLFKK905 pKa = 11.17NLLSTDD911 pKa = 3.69AVHH914 pKa = 6.96KK915 pKa = 10.21GSKK918 pKa = 9.22FVWYY922 pKa = 8.18TFNPKK927 pKa = 9.74IPGHH931 pKa = 5.78IASMARR937 pKa = 11.84RR938 pKa = 11.84FLNLVTFSSDD948 pKa = 2.86KK949 pKa = 9.69EE950 pKa = 4.4TWDD953 pKa = 3.22LSVRR957 pKa = 11.84IEE959 pKa = 4.49FYY961 pKa = 11.18RR962 pKa = 11.84MLARR966 pKa = 11.84IVQNLCKK973 pKa = 9.8KK974 pKa = 9.51HH975 pKa = 6.6PDD977 pKa = 3.47MTVEE981 pKa = 4.33AYY983 pKa = 9.68KK984 pKa = 10.5QVLLDD989 pKa = 3.49IYY991 pKa = 9.79DD992 pKa = 4.29TIMQVGAEE1000 pKa = 4.08GEE1002 pKa = 4.12YY1003 pKa = 10.62RR1004 pKa = 11.84LNNGYY1009 pKa = 7.85VLKK1012 pKa = 10.6AVYY1015 pKa = 9.95AGYY1018 pKa = 10.39LKK1020 pKa = 10.74SGWYY1024 pKa = 8.07LTIIANSIMQVVVHH1038 pKa = 5.91VLACFRR1044 pKa = 11.84CGMSFEE1050 pKa = 4.96EE1051 pKa = 5.5IIDD1054 pKa = 3.87LPFVAGGDD1062 pKa = 3.81DD1063 pKa = 4.49VIQHH1067 pKa = 6.49LPDD1070 pKa = 5.19HH1071 pKa = 6.43IVDD1074 pKa = 4.53RR1075 pKa = 11.84YY1076 pKa = 10.28IEE1078 pKa = 3.91AHH1080 pKa = 6.0ASFGIKK1086 pKa = 10.06LKK1088 pKa = 10.81LEE1090 pKa = 3.83RR1091 pKa = 11.84HH1092 pKa = 5.46EE1093 pKa = 4.35SFEE1096 pKa = 3.93GSEE1099 pKa = 4.19FFSNRR1104 pKa = 11.84FFLDD1108 pKa = 4.08SKK1110 pKa = 9.87GKK1112 pKa = 9.32WYY1114 pKa = 9.85FVPQRR1119 pKa = 11.84FCKK1122 pKa = 10.33HH1123 pKa = 5.78IEE1125 pKa = 4.22SLLHH1129 pKa = 5.96MKK1131 pKa = 10.36PKK1133 pKa = 10.74DD1134 pKa = 3.56LFQGLSSKK1142 pKa = 11.02LDD1144 pKa = 3.25DD1145 pKa = 3.43WCFKK1149 pKa = 11.34DD1150 pKa = 4.14EE1151 pKa = 4.09VSQVFEE1157 pKa = 5.83DD1158 pKa = 3.26IMSVIRR1164 pKa = 11.84TKK1166 pKa = 10.82YY1167 pKa = 8.0KK1168 pKa = 9.71HH1169 pKa = 6.55LYY1171 pKa = 8.77VPSVDD1176 pKa = 3.0KK1177 pKa = 10.68TRR1179 pKa = 11.84RR1180 pKa = 11.84EE1181 pKa = 4.56AINQARR1187 pKa = 11.84GYY1189 pKa = 10.56EE1190 pKa = 3.99DD1191 pKa = 4.89LPINAEE1197 pKa = 3.88TAIACDD1203 pKa = 3.54SQTQTDD1209 pKa = 3.99EE1210 pKa = 4.2KK1211 pKa = 11.31VEE1213 pKa = 4.46DD1214 pKa = 4.18GGDD1217 pKa = 3.52FRR1219 pKa = 11.84AEE1221 pKa = 3.56FEE1223 pKa = 4.52AIKK1226 pKa = 10.84KK1227 pKa = 8.69EE1228 pKa = 4.06MRR1230 pKa = 11.84QMMHH1234 pKa = 6.47TYY1236 pKa = 10.62RR1237 pKa = 11.84MQIDD1241 pKa = 3.93ALTSTKK1247 pKa = 10.85NKK1249 pKa = 9.14LQKK1252 pKa = 10.28KK1253 pKa = 8.38VLHH1256 pKa = 6.77LEE1258 pKa = 4.37TQLVTAKK1265 pKa = 10.29LKK1267 pKa = 11.19NKK1269 pKa = 9.63GVSNNIGKK1277 pKa = 9.9RR1278 pKa = 11.84KK1279 pKa = 9.77KK1280 pKa = 10.18DD1281 pKa = 3.64VNSQSSDD1288 pKa = 3.22PVLRR1292 pKa = 11.84SPKK1295 pKa = 9.86SPVGATVPTVDD1306 pKa = 4.98QVVQPQSGGEE1316 pKa = 4.07VFPPLPRR1323 pKa = 11.84DD1324 pKa = 3.56CC1325 pKa = 5.86
MM1 pKa = 7.6IVPKK5 pKa = 10.85NKK7 pKa = 9.32FCQSDD12 pKa = 3.71DD13 pKa = 3.41YY14 pKa = 11.57DD15 pKa = 3.58YY16 pKa = 11.52CSRR19 pKa = 11.84DD20 pKa = 3.46KK21 pKa = 11.17CWVCLIPLRR30 pKa = 11.84CSDD33 pKa = 5.26LIEE36 pKa = 5.01KK37 pKa = 9.66YY38 pKa = 10.14ISYY41 pKa = 10.61DD42 pKa = 3.08PYY44 pKa = 10.38EE45 pKa = 4.56AFGSEE50 pKa = 4.47YY51 pKa = 10.62EE52 pKa = 4.63SIVLPCEE59 pKa = 3.58RR60 pKa = 11.84LAALCYY66 pKa = 10.45YY67 pKa = 9.99EE68 pKa = 5.11SLWTEE73 pKa = 3.9VRR75 pKa = 11.84RR76 pKa = 11.84SSTEE80 pKa = 3.24WSIARR85 pKa = 11.84WYY87 pKa = 8.62STAVDD92 pKa = 3.21HH93 pKa = 7.26RR94 pKa = 11.84GRR96 pKa = 11.84PRR98 pKa = 11.84AFRR101 pKa = 11.84IVGTNLHH108 pKa = 6.77DD109 pKa = 4.27LAGVAAMPYY118 pKa = 8.65PPTMSEE124 pKa = 3.58MEE126 pKa = 4.62YY127 pKa = 10.2ILKK130 pKa = 9.81EE131 pKa = 4.21VPHH134 pKa = 7.29DD135 pKa = 4.98DD136 pKa = 3.7IPGEE140 pKa = 4.08VFKK143 pKa = 11.39YY144 pKa = 10.49LGGRR148 pKa = 11.84TKK150 pKa = 11.02LSFTLKK156 pKa = 10.05LFLMFGVLSGTIFSVEE172 pKa = 4.05ASDD175 pKa = 5.17QYY177 pKa = 11.61TSDD180 pKa = 3.43LVNTGEE186 pKa = 4.23LARR189 pKa = 11.84EE190 pKa = 3.78ITLWPVNEE198 pKa = 4.15SFRR201 pKa = 11.84MLEE204 pKa = 3.87HH205 pKa = 5.76TTISTTQCHH214 pKa = 5.8YY215 pKa = 11.48EE216 pKa = 3.85NGTCVEE222 pKa = 4.23YY223 pKa = 10.41PNLVNQVFDD232 pKa = 3.62WLRR235 pKa = 11.84VRR237 pKa = 11.84VRR239 pKa = 11.84NLCYY243 pKa = 10.63LLDD246 pKa = 4.23EE247 pKa = 4.54STLLARR253 pKa = 11.84HH254 pKa = 5.95SSSVAKK260 pKa = 10.26EE261 pKa = 3.37RR262 pKa = 11.84TRR264 pKa = 11.84FYY266 pKa = 11.47KK267 pKa = 10.71NIIVKK272 pKa = 10.22DD273 pKa = 3.79DD274 pKa = 3.66YY275 pKa = 11.47FRR277 pKa = 11.84AEE279 pKa = 4.09YY280 pKa = 10.89DD281 pKa = 4.21DD282 pKa = 5.07ISTAEE287 pKa = 3.89NEE289 pKa = 4.75LYY291 pKa = 11.03NFMMLFLSFLYY302 pKa = 10.42GINSGFWSLCILFINFLYY320 pKa = 10.41FMRR323 pKa = 11.84FAHH326 pKa = 6.02NFVVSTPLGFWIYY339 pKa = 10.54TKK341 pKa = 10.19VVWVLGIWLGYY352 pKa = 8.78YY353 pKa = 8.12VTYY356 pKa = 10.15HH357 pKa = 6.59ALVVLLAASISYY369 pKa = 9.23LHH371 pKa = 6.72SKK373 pKa = 9.63MIEE376 pKa = 3.94LAVVVVMAPYY386 pKa = 9.94HH387 pKa = 4.78IWSRR391 pKa = 11.84FRR393 pKa = 11.84RR394 pKa = 11.84TGVKK398 pKa = 10.04GFVRR402 pKa = 11.84EE403 pKa = 4.13MILPGSKK410 pKa = 8.86IEE412 pKa = 4.16KK413 pKa = 10.2FIGAKK418 pKa = 9.31QPAVAALVVEE428 pKa = 4.94VDD430 pKa = 4.22GKK432 pKa = 7.88WHH434 pKa = 5.8YY435 pKa = 11.43SAMCFRR441 pKa = 11.84YY442 pKa = 9.86EE443 pKa = 3.85NHH445 pKa = 6.48LVSAFHH451 pKa = 7.01PFAKK455 pKa = 10.12HH456 pKa = 4.43MSVGRR461 pKa = 11.84RR462 pKa = 11.84MGVAPNLLCKK472 pKa = 10.33KK473 pKa = 9.83RR474 pKa = 11.84WTHH477 pKa = 6.81DD478 pKa = 3.32FGAIQVIDD486 pKa = 3.97PSNYY490 pKa = 7.98IVDD493 pKa = 4.53FEE495 pKa = 4.34RR496 pKa = 11.84HH497 pKa = 5.58DD498 pKa = 3.7VFMLHH503 pKa = 6.27MSGEE507 pKa = 3.78FWDD510 pKa = 4.01RR511 pKa = 11.84VRR513 pKa = 11.84LPEE516 pKa = 4.18PKK518 pKa = 10.07KK519 pKa = 10.74SVSNSFNDD527 pKa = 3.32SATVFSYY534 pKa = 10.17MAGTLHH540 pKa = 6.6TSNGVLTSHH549 pKa = 6.78TNVDD553 pKa = 3.66YY554 pKa = 10.68PPNNFMAHH562 pKa = 5.43TCSTLPGFSGTPVFAANGAVRR583 pKa = 11.84GVHH586 pKa = 6.11IGSDD590 pKa = 3.33NYY592 pKa = 10.31EE593 pKa = 3.83EE594 pKa = 4.36RR595 pKa = 11.84NIMFRR600 pKa = 11.84IDD602 pKa = 3.45HH603 pKa = 6.32IKK605 pKa = 10.93YY606 pKa = 9.55YY607 pKa = 10.76LNKK610 pKa = 10.43NKK612 pKa = 8.07VTRR615 pKa = 11.84EE616 pKa = 3.98DD617 pKa = 3.35SSDD620 pKa = 3.56FQKK623 pKa = 11.08KK624 pKa = 7.11VLRR627 pKa = 11.84SMKK630 pKa = 9.64RR631 pKa = 11.84RR632 pKa = 11.84KK633 pKa = 9.86GKK635 pKa = 9.17EE636 pKa = 3.24AVYY639 pKa = 10.9YY640 pKa = 10.73FDD642 pKa = 6.44DD643 pKa = 4.25FDD645 pKa = 4.56EE646 pKa = 5.43DD647 pKa = 3.56NSVAYY652 pKa = 9.86DD653 pKa = 3.22IRR655 pKa = 11.84TGQIEE660 pKa = 4.27RR661 pKa = 11.84VPRR664 pKa = 11.84TEE666 pKa = 4.36EE667 pKa = 3.54EE668 pKa = 4.67DD669 pKa = 3.6EE670 pKa = 4.32EE671 pKa = 4.37FWRR674 pKa = 11.84VEE676 pKa = 3.71MQKK679 pKa = 10.5PVGLWGDD686 pKa = 3.84YY687 pKa = 10.65EE688 pKa = 4.26DD689 pKa = 5.47APISKK694 pKa = 10.09EE695 pKa = 3.94EE696 pKa = 4.51EE697 pKa = 3.73IDD699 pKa = 3.65EE700 pKa = 4.39VEE702 pKa = 3.9PSQFFEE708 pKa = 4.17VLDD711 pKa = 3.73MRR713 pKa = 11.84KK714 pKa = 9.37KK715 pKa = 10.31LHH717 pKa = 6.71SPTPDD722 pKa = 2.93TKK724 pKa = 10.65HH725 pKa = 6.98AEE727 pKa = 4.08TTEE730 pKa = 3.76ILEE733 pKa = 4.17PFMDD737 pKa = 3.63KK738 pKa = 10.56FKK740 pKa = 10.99EE741 pKa = 3.92LGYY744 pKa = 10.13VEE746 pKa = 6.25GMFEE750 pKa = 4.06YY751 pKa = 9.84PAAGHH756 pKa = 6.93KK757 pKa = 9.42MDD759 pKa = 4.61TKK761 pKa = 10.85SCVNAIIQHH770 pKa = 4.94GTNVSEE776 pKa = 4.85CPVPITPDD784 pKa = 3.24EE785 pKa = 4.14EE786 pKa = 4.54DD787 pKa = 3.41RR788 pKa = 11.84VVDD791 pKa = 3.81AVSAMVQEE799 pKa = 4.87FSFVVDD805 pKa = 3.87PNYY808 pKa = 10.83KK809 pKa = 9.1SLEE812 pKa = 4.02YY813 pKa = 10.55FEE815 pKa = 6.44SIINTNQIEE824 pKa = 4.27DD825 pKa = 3.72RR826 pKa = 11.84KK827 pKa = 10.24SPGMPYY833 pKa = 10.21LEE835 pKa = 4.51EE836 pKa = 5.27GMTTNAQVLDD846 pKa = 3.9KK847 pKa = 11.51LGVTWLYY854 pKa = 11.74NKK856 pKa = 9.83MIEE859 pKa = 4.29CWNMSILYY867 pKa = 10.42RR868 pKa = 11.84MFLKK872 pKa = 10.98YY873 pKa = 10.63EE874 pKa = 3.8PTKK877 pKa = 10.55KK878 pKa = 10.43KK879 pKa = 10.83KK880 pKa = 9.31IDD882 pKa = 2.84NWMARR887 pKa = 11.84VILSFPLHH895 pKa = 6.0KK896 pKa = 9.46TLQNIVLFKK905 pKa = 11.17NLLSTDD911 pKa = 3.69AVHH914 pKa = 6.96KK915 pKa = 10.21GSKK918 pKa = 9.22FVWYY922 pKa = 8.18TFNPKK927 pKa = 9.74IPGHH931 pKa = 5.78IASMARR937 pKa = 11.84RR938 pKa = 11.84FLNLVTFSSDD948 pKa = 2.86KK949 pKa = 9.69EE950 pKa = 4.4TWDD953 pKa = 3.22LSVRR957 pKa = 11.84IEE959 pKa = 4.49FYY961 pKa = 11.18RR962 pKa = 11.84MLARR966 pKa = 11.84IVQNLCKK973 pKa = 9.8KK974 pKa = 9.51HH975 pKa = 6.6PDD977 pKa = 3.47MTVEE981 pKa = 4.33AYY983 pKa = 9.68KK984 pKa = 10.5QVLLDD989 pKa = 3.49IYY991 pKa = 9.79DD992 pKa = 4.29TIMQVGAEE1000 pKa = 4.08GEE1002 pKa = 4.12YY1003 pKa = 10.62RR1004 pKa = 11.84LNNGYY1009 pKa = 7.85VLKK1012 pKa = 10.6AVYY1015 pKa = 9.95AGYY1018 pKa = 10.39LKK1020 pKa = 10.74SGWYY1024 pKa = 8.07LTIIANSIMQVVVHH1038 pKa = 5.91VLACFRR1044 pKa = 11.84CGMSFEE1050 pKa = 4.96EE1051 pKa = 5.5IIDD1054 pKa = 3.87LPFVAGGDD1062 pKa = 3.81DD1063 pKa = 4.49VIQHH1067 pKa = 6.49LPDD1070 pKa = 5.19HH1071 pKa = 6.43IVDD1074 pKa = 4.53RR1075 pKa = 11.84YY1076 pKa = 10.28IEE1078 pKa = 3.91AHH1080 pKa = 6.0ASFGIKK1086 pKa = 10.06LKK1088 pKa = 10.81LEE1090 pKa = 3.83RR1091 pKa = 11.84HH1092 pKa = 5.46EE1093 pKa = 4.35SFEE1096 pKa = 3.93GSEE1099 pKa = 4.19FFSNRR1104 pKa = 11.84FFLDD1108 pKa = 4.08SKK1110 pKa = 9.87GKK1112 pKa = 9.32WYY1114 pKa = 9.85FVPQRR1119 pKa = 11.84FCKK1122 pKa = 10.33HH1123 pKa = 5.78IEE1125 pKa = 4.22SLLHH1129 pKa = 5.96MKK1131 pKa = 10.36PKK1133 pKa = 10.74DD1134 pKa = 3.56LFQGLSSKK1142 pKa = 11.02LDD1144 pKa = 3.25DD1145 pKa = 3.43WCFKK1149 pKa = 11.34DD1150 pKa = 4.14EE1151 pKa = 4.09VSQVFEE1157 pKa = 5.83DD1158 pKa = 3.26IMSVIRR1164 pKa = 11.84TKK1166 pKa = 10.82YY1167 pKa = 8.0KK1168 pKa = 9.71HH1169 pKa = 6.55LYY1171 pKa = 8.77VPSVDD1176 pKa = 3.0KK1177 pKa = 10.68TRR1179 pKa = 11.84RR1180 pKa = 11.84EE1181 pKa = 4.56AINQARR1187 pKa = 11.84GYY1189 pKa = 10.56EE1190 pKa = 3.99DD1191 pKa = 4.89LPINAEE1197 pKa = 3.88TAIACDD1203 pKa = 3.54SQTQTDD1209 pKa = 3.99EE1210 pKa = 4.2KK1211 pKa = 11.31VEE1213 pKa = 4.46DD1214 pKa = 4.18GGDD1217 pKa = 3.52FRR1219 pKa = 11.84AEE1221 pKa = 3.56FEE1223 pKa = 4.52AIKK1226 pKa = 10.84KK1227 pKa = 8.69EE1228 pKa = 4.06MRR1230 pKa = 11.84QMMHH1234 pKa = 6.47TYY1236 pKa = 10.62RR1237 pKa = 11.84MQIDD1241 pKa = 3.93ALTSTKK1247 pKa = 10.85NKK1249 pKa = 9.14LQKK1252 pKa = 10.28KK1253 pKa = 8.38VLHH1256 pKa = 6.77LEE1258 pKa = 4.37TQLVTAKK1265 pKa = 10.29LKK1267 pKa = 11.19NKK1269 pKa = 9.63GVSNNIGKK1277 pKa = 9.9RR1278 pKa = 11.84KK1279 pKa = 9.77KK1280 pKa = 10.18DD1281 pKa = 3.64VNSQSSDD1288 pKa = 3.22PVLRR1292 pKa = 11.84SPKK1295 pKa = 9.86SPVGATVPTVDD1306 pKa = 4.98QVVQPQSGGEE1316 pKa = 4.07VFPPLPRR1323 pKa = 11.84DD1324 pKa = 3.56CC1325 pKa = 5.86
Molecular weight: 153.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KED1|A0A1L3KED1_9VIRU RNA-directed RNA polymerase OS=Changjiang sobemo-like virus 2 OX=1922801 PE=4 SV=1
MM1 pKa = 7.22YY2 pKa = 10.38KK3 pKa = 10.45LHH5 pKa = 6.35VKK7 pKa = 10.56NNGLDD12 pKa = 3.49TTIFNPNCSSEE23 pKa = 4.09NLMSSARR30 pKa = 11.84EE31 pKa = 3.62QKK33 pKa = 10.09QNKK36 pKa = 8.32KK37 pKa = 9.65NAYY40 pKa = 8.14PDD42 pKa = 3.97EE43 pKa = 4.53YY44 pKa = 11.38DD45 pKa = 3.82NLPDD49 pKa = 3.79SSFPYY54 pKa = 9.66FDD56 pKa = 5.16NYY58 pKa = 9.58TGPYY62 pKa = 8.18WSDD65 pKa = 2.84GKK67 pKa = 10.79FQSSVNNGQSKK78 pKa = 9.74PLSKK82 pKa = 10.65LDD84 pKa = 3.19WHH86 pKa = 6.85SRR88 pKa = 11.84DD89 pKa = 3.65HH90 pKa = 6.93DD91 pKa = 3.7ACFADD96 pKa = 4.49GKK98 pKa = 10.69DD99 pKa = 2.89IAYY102 pKa = 10.52CDD104 pKa = 3.36DD105 pKa = 3.4VYY107 pKa = 11.87YY108 pKa = 11.43NNTRR112 pKa = 11.84DD113 pKa = 3.26LDD115 pKa = 3.74LRR117 pKa = 11.84GRR119 pKa = 11.84LIGIIPKK126 pKa = 9.47LAHH129 pKa = 6.34GGFSIFGQSGRR140 pKa = 11.84VGGVADD146 pKa = 3.85KK147 pKa = 7.77MTRR150 pKa = 11.84PTPAQSAGGHH160 pKa = 5.87PSGPTSYY167 pKa = 10.82SGVPTDD173 pKa = 3.33SLKK176 pKa = 10.95PIGGVYY182 pKa = 10.49DD183 pKa = 4.5GIGLVAKK190 pKa = 10.08PSMPNSFPKK199 pKa = 10.64HH200 pKa = 5.86PDD202 pKa = 3.09FGEE205 pKa = 4.2STLLQTTYY213 pKa = 11.1APEE216 pKa = 4.03TQEE219 pKa = 3.84PKK221 pKa = 10.85VIDD224 pKa = 3.67RR225 pKa = 11.84QEE227 pKa = 3.97LSEE230 pKa = 4.94NPLNLNPTRR239 pKa = 11.84DD240 pKa = 3.76DD241 pKa = 3.62QVGYY245 pKa = 8.65TYY247 pKa = 10.65EE248 pKa = 3.96SKK250 pKa = 10.88GRR252 pKa = 11.84YY253 pKa = 8.65NSQKK257 pKa = 8.27TWQKK261 pKa = 9.7PRR263 pKa = 11.84KK264 pKa = 9.2HH265 pKa = 5.95KK266 pKa = 9.83NQRR269 pKa = 11.84KK270 pKa = 8.87RR271 pKa = 11.84RR272 pKa = 11.84NKK274 pKa = 10.16RR275 pKa = 11.84KK276 pKa = 9.68NKK278 pKa = 9.61KK279 pKa = 9.49
MM1 pKa = 7.22YY2 pKa = 10.38KK3 pKa = 10.45LHH5 pKa = 6.35VKK7 pKa = 10.56NNGLDD12 pKa = 3.49TTIFNPNCSSEE23 pKa = 4.09NLMSSARR30 pKa = 11.84EE31 pKa = 3.62QKK33 pKa = 10.09QNKK36 pKa = 8.32KK37 pKa = 9.65NAYY40 pKa = 8.14PDD42 pKa = 3.97EE43 pKa = 4.53YY44 pKa = 11.38DD45 pKa = 3.82NLPDD49 pKa = 3.79SSFPYY54 pKa = 9.66FDD56 pKa = 5.16NYY58 pKa = 9.58TGPYY62 pKa = 8.18WSDD65 pKa = 2.84GKK67 pKa = 10.79FQSSVNNGQSKK78 pKa = 9.74PLSKK82 pKa = 10.65LDD84 pKa = 3.19WHH86 pKa = 6.85SRR88 pKa = 11.84DD89 pKa = 3.65HH90 pKa = 6.93DD91 pKa = 3.7ACFADD96 pKa = 4.49GKK98 pKa = 10.69DD99 pKa = 2.89IAYY102 pKa = 10.52CDD104 pKa = 3.36DD105 pKa = 3.4VYY107 pKa = 11.87YY108 pKa = 11.43NNTRR112 pKa = 11.84DD113 pKa = 3.26LDD115 pKa = 3.74LRR117 pKa = 11.84GRR119 pKa = 11.84LIGIIPKK126 pKa = 9.47LAHH129 pKa = 6.34GGFSIFGQSGRR140 pKa = 11.84VGGVADD146 pKa = 3.85KK147 pKa = 7.77MTRR150 pKa = 11.84PTPAQSAGGHH160 pKa = 5.87PSGPTSYY167 pKa = 10.82SGVPTDD173 pKa = 3.33SLKK176 pKa = 10.95PIGGVYY182 pKa = 10.49DD183 pKa = 4.5GIGLVAKK190 pKa = 10.08PSMPNSFPKK199 pKa = 10.64HH200 pKa = 5.86PDD202 pKa = 3.09FGEE205 pKa = 4.2STLLQTTYY213 pKa = 11.1APEE216 pKa = 4.03TQEE219 pKa = 3.84PKK221 pKa = 10.85VIDD224 pKa = 3.67RR225 pKa = 11.84QEE227 pKa = 3.97LSEE230 pKa = 4.94NPLNLNPTRR239 pKa = 11.84DD240 pKa = 3.76DD241 pKa = 3.62QVGYY245 pKa = 8.65TYY247 pKa = 10.65EE248 pKa = 3.96SKK250 pKa = 10.88GRR252 pKa = 11.84YY253 pKa = 8.65NSQKK257 pKa = 8.27TWQKK261 pKa = 9.7PRR263 pKa = 11.84KK264 pKa = 9.2HH265 pKa = 5.95KK266 pKa = 9.83NQRR269 pKa = 11.84KK270 pKa = 8.87RR271 pKa = 11.84RR272 pKa = 11.84NKK274 pKa = 10.16RR275 pKa = 11.84KK276 pKa = 9.68NKK278 pKa = 9.61KK279 pKa = 9.49
Molecular weight: 31.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2001 |
279 |
1325 |
667.0 |
75.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.947 ± 1.177 | 1.549 ± 0.265 |
5.897 ± 0.646 | 5.297 ± 1.732 |
4.948 ± 0.75 | 5.847 ± 1.049 |
2.549 ± 0.516 | 5.097 ± 0.588 |
6.547 ± 0.766 | 7.296 ± 0.57 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.849 ± 0.586 | 4.798 ± 0.911 |
5.247 ± 0.983 | 3.198 ± 0.659 |
4.748 ± 0.626 | 7.996 ± 0.991 |
5.797 ± 0.889 | 7.796 ± 1.248 |
1.849 ± 0.274 | 4.748 ± 0.209 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
